Expansion of a unique region in the Marek's disease virus genome occurs concomitantly with attenuation but is not sufficient to cause attenuation
- PMID: 14694105
- PMCID: PMC368850
- DOI: 10.1128/jvi.78.2.733-740.2004
Expansion of a unique region in the Marek's disease virus genome occurs concomitantly with attenuation but is not sufficient to cause attenuation
Abstract
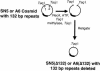
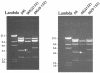
Pathogenic Marek's disease viruses (MDVs) have two head-to-tail copies of a 132-bp repeat. As MDV is serially passaged in cell culture, the virus becomes attenuated and the number of copies of the 132-bp repeat increases from 2 to often more than 20 copies. To determine the role of the repeats in attenuation, we used five overlapping cosmid clones that spanned the MDV genome to reconstitute infectious virus (rMd5). By mutating the appropriate cosmids, we generated clones of infectious MDVs that contained zero copies of the 132-bp repeats, rMd5(Delta132); nine copies of the 132-bp repeats, rMd5(9-132); and nine copies of the 132-bp repeats inserted in the reverse orientation, rMd5(rev9-132). After two passages in cell culture, wild-type Md5, rMd5, and rMd5(Delta132) were stable. However, rMd5(9-132) and rMd5(rev9-132) contained a population of viruses that contained from 3 to over 20 copies of the repeats. A major 1.8-kb mRNA, containing two copies of the 132-bp repeat, was present in wild-type Md5 and rMd5 but was not present in rMd5(Delta132), rMd5(9-132), rMd5(rev9-132), or an attenuated MDV. Instead, the RNAs transcribed from the 132-bp repeat region in rMd5(9-132) and rMd5(rev9-132) closely resembled the pattern of RNAs transcribed in attenuated MDVs. When inoculated into susceptible day-old chicks, all viruses produced various lesions. Thus, expansion of the number of copies of 132-bp repeats, which accompanies attenuation, is not sufficient in itself to attenuate pathogenic MDVs.
Figures


Similar articles
-
Marek's Disease Virus (MDV) Meq Oncoprotein Plays Distinct Roles in Tumor Incidence, Distribution, and Size.Viruses. 2025 Feb 14;17(2):259. doi: 10.3390/v17020259. Viruses. 2025. PMID: 40007015 Free PMC article.
-
Marek's disease virus-encoded vIL-8 gene is involved in early cytolytic infection but dispensable for establishment of latency.J Virol. 2004 May;78(9):4753-60. doi: 10.1128/jvi.78.9.4753-4760.2004. J Virol. 2004. PMID: 15078957 Free PMC article.
-
Marek's Disease Virus Requires Both Copies of the Inverted Repeat Regions for Efficient In Vivo Replication and Pathogenesis.J Virol. 2021 Jan 13;95(3):e01256-20. doi: 10.1128/JVI.01256-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33115875 Free PMC article.
-
Marek's disease virus and skin interactions.Vet Res. 2014 Apr 3;45(1):36. doi: 10.1186/1297-9716-45-36. Vet Res. 2014. PMID: 24694064 Free PMC article. Review.
-
Latency and tumorigenesis in Marek's disease.Avian Dis. 2013 Jun;57(2 Suppl):360-5. doi: 10.1637/10470-121712-Reg.1. Avian Dis. 2013. PMID: 23901747 Review.
Cited by
-
Isolation and purification of Gallid herpesvirus 2 strains currently distributed in Japan.J Vet Med Sci. 2017 Jan 20;79(1):115-122. doi: 10.1292/jvms.16-0329. Epub 2016 Oct 8. J Vet Med Sci. 2017. PMID: 27725354 Free PMC article.
-
Viral proteogenomic and expression profiling during productive replication of a skin-tropic herpesvirus in the natural host.PLoS Pathog. 2023 Jun 8;19(6):e1011204. doi: 10.1371/journal.ppat.1011204. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37289833 Free PMC article.
-
The role of pp38 in regulation of Marek's disease virus bi-directional promoter between pp38 and 1.8-kb mRNA.Virus Genes. 2006 Apr;32(2):193-201. doi: 10.1007/s11262-005-6876-2. Virus Genes. 2006. PMID: 16604452
-
Sequence determination of a mildly virulent strain (CU-2) of Gallid herpesvirus type 2 using 454 pyrosequencing.Virus Genes. 2008 Jun;36(3):479-89. doi: 10.1007/s11262-008-0213-5. Epub 2008 Mar 20. Virus Genes. 2008. PMID: 18351449
-
Oncogenic Marek's disease viruses lacking the 132 base pair repeats can still be attenuated by serial in vitro cell culture passages.Virus Genes. 2007 Jan;34(1):87-90. doi: 10.1007/s11262-006-0022-7. Epub 2006 Aug 22. Virus Genes. 2007. PMID: 16927124
References
-
- Bülow, V. V., and P. M. Biggs. 1975. Precipitating antigens associated with Marek's disease virus and a herpesvirus of turkeys. Avian Pathol. 4:147-162. - PubMed
-
- Chomczynski, P., and N. Sacchi. 1987. Single-step method RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

