Functional analysis of the human papillomavirus type 16 E1=E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization
- PMID: 14694114
- PMCID: PMC368840
- DOI: 10.1128/jvi.78.2.821-833.2004
Functional analysis of the human papillomavirus type 16 E1=E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization
Abstract
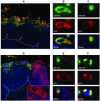
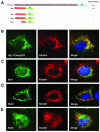
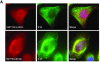
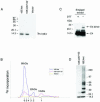
High-risk human papillomaviruses, such as human papillomavirus type 16 (HPV16), are the primary cause of cervical cancer. The HPV16 E1=E4 protein associates with keratin intermediate filaments and causes network collapse when expressed in epithelial cells in vitro. Here, we show that keratin association and network reorganization also occur in vivo in low-grade cervical neoplasia caused by HPV16. The 16E1=E4 protein binds to keratins directly and interacts strongly with keratin 18, a member of the type I intermediate-filament family. By contrast, 16E1=E4 bound only weakly to keratin 8, a type II intermediate-filament protein, and showed no detectable affinity for the type III protein, vimentin. The N-terminal 16 amino acids of the 16E1=E4 protein, which contains the YPLLXLL motif that is conserved among supergroup A viruses, were sufficient to target green fluorescent protein to the keratin network. When expressed in the SiHa cervical epithelial cell line, the full-length 16E1=E4 protein caused an almost total inhibition of keratin dynamics, despite the phosphorylation of keratin 18 at serine 33, which normally leads to 14-3-3-mediated keratin solubilization. Mutant 16E1=E4 proteins which lack the LLKLL motif, or which have lost amino acids from their C termini, and which were compromised in the ability to associate with keratins did not disturb normal keratin dynamics. 16E1=E4 was found to exist as dimers and hexamers, whereas a C-terminal deletion mutant (16E1=E4Delta87-92) existed as monomers and formed multimeric structures only poorly. Considered together, our results suggest that by associating with keratins through its N terminus, and by associating with itself through its C terminus, 16E1=E4 may act as a keratin cross-linker and prevent the movement of keratins between the soluble and insoluble compartments. The increase in avidity associated with multimeric binding may contribute to the ability of 16E1=E4 to sequester its cellular targets in the cytoplasm.
Figures



Similar articles
-
Phosphorylation of the human papillomavirus type 16 E1--E4 protein at T57 by ERK triggers a structural change that enhances keratin binding and protein stability.J Virol. 2009 Apr;83(8):3668-83. doi: 10.1128/JVI.02063-08. Epub 2009 Feb 11. J Virol. 2009. PMID: 19211765 Free PMC article.
-
Identification of a G(2) arrest domain in the E1 wedge E4 protein of human papillomavirus type 16.J Virol. 2002 Oct;76(19):9806-18. doi: 10.1128/jvi.76.19.9806-9818.2002. J Virol. 2002. PMID: 12208959 Free PMC article.
-
Role of calpain in the formation of human papillomavirus type 16 E1^E4 amyloid fibers and reorganization of the keratin network.J Virol. 2011 Oct;85(19):9984-97. doi: 10.1128/JVI.02158-10. Epub 2011 Jul 13. J Virol. 2011. PMID: 21752901 Free PMC article.
-
Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia.J Cell Biol. 2011 Sep 5;194(5):669-78. doi: 10.1083/jcb.201008095. J Cell Biol. 2011. PMID: 21893596 Free PMC article. Review.
-
Keratin modifications and solubility properties in epithelial cells and in vitro.Subcell Biochem. 1998;31:105-40. Subcell Biochem. 1998. PMID: 9932491 Review.
Cited by
-
Tissue-spanning redox gradient-dependent assembly of native human papillomavirus type 16 virions.J Virol. 2009 Oct;83(20):10515-26. doi: 10.1128/JVI.00731-09. Epub 2009 Aug 5. J Virol. 2009. PMID: 19656879 Free PMC article.
-
HPV16 and 18 genome amplification show different E4-dependence, with 16E4 enhancing E1 nuclear accumulation and replicative efficiency via its cell cycle arrest and kinase activation functions.PLoS Pathog. 2017 Mar 17;13(3):e1006282. doi: 10.1371/journal.ppat.1006282. eCollection 2017 Mar. PLoS Pathog. 2017. PMID: 28306742 Free PMC article.
-
Human papillomavirus 18 E1^E4 protein interacts with cyclin A/CDK 2 through an RXL motif.Mol Cell Biochem. 2013 Jan;373(1-2):29-40. doi: 10.1007/s11010-012-1472-y. Epub 2012 Oct 13. Mol Cell Biochem. 2013. PMID: 23065011
-
Phosphorylation and Reorganization of Keratin Networks: Implications for Carcinogenesis and Epithelial Mesenchymal Transition.Biomol Ther (Seoul). 2015 Jul;23(4):301-12. doi: 10.4062/biomolther.2015.032. Epub 2015 Jul 1. Biomol Ther (Seoul). 2015. PMID: 26157545 Free PMC article. Review.
-
Human papillomavirus type 16 E5 protein as a therapeutic target.Yonsei Med J. 2006 Feb 28;47(1):1-14. doi: 10.3349/ymj.2006.47.1.1. Yonsei Med J. 2006. Retraction in: Yonsei Med J. 2011 May;52(3):551. doi: 10.3349/ymj.2011.52.3.551. PMID: 16502480 Free PMC article. Retracted. Review.
References
-
- Ashmole, I., P. H. Gallimore, and S. Roberts. 1998. Identification of conserved hydrophobic C-terminal residues of the human papillomavirus type 1 E1∧E4 protein necessary for E4 oligomerisation in vivo. Virology 240:221-231. - PubMed
-
- Bryan, J. T., K. H. Fife, and D. R. Brown. 1998. The intracellular expression pattern of the human papillomavirus type 11 E1∧E4 protein correlates with its ability to self associate. Virology 241:49-60. - PubMed
-
- Chou, C. F., C. L. Riopel, L. S. Rott, and M. B. Omary. 1993. A significant soluble keratin fraction in ‘simple' epithelial cells. Lack of an apparent phosphorylation and glycosylation role in keratin solubility. J. Cell Sci. 105:433-444. - PubMed
-
- Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

