A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts
- PMID: 14694196
- PMCID: PMC314136
- DOI: 10.1073/pnas.0307163101
A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts
Abstract
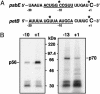
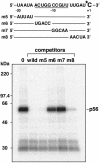
RNA editing involves a variety of genetic systems and occurs by different mechanisms. In higher plant chloroplasts, specific sites of some transcripts are subject to C-to-U conversion. We have previously shown that site-specific trans-acting factors for psbE and petB mRNA editing bind corresponding cis-elements, which are located 5 nucleotides upstream from the editing site. Here we report that, by using mRNAs labeled either at the center of the upstream cis-element or at the editing site, the site-specific factors can be cross-linked with nucleotides at both positions. Mutations of nucleotides in the proximal region of the editing site revealed a correlation between editing activity and cross-linking efficiency of factors with the editing site, even though cross-linking with the upstream cis-element was unaffected. These observations suggest that the site-specific factor binds stably to the upstream ciselement, whereas it interacts weakly with the editing site. This finding raises the intriguing possibility that the site-specific factor is involved in both site-determination and C-to-U conversion in chloroplast RNA editing.
Figures




Similar articles
-
A simple in vitro RNA editing assay for chloroplast transcripts using fluorescent dideoxynucleotides: distinct types of sequence elements required for editing of ndh transcripts.Plant J. 2006 Sep;47(5):802-10. doi: 10.1111/j.1365-313X.2006.02825.x. Epub 2006 Jul 11. Plant J. 2006. PMID: 16856984
-
Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system.EMBO J. 2001 Mar 1;20(5):1144-52. doi: 10.1093/emboj/20.5.1144. EMBO J. 2001. PMID: 11230137 Free PMC article.
-
Identification of critical nucleotide positions for plastid RNA editing site recognition.RNA. 1997 Oct;3(10):1194-200. RNA. 1997. PMID: 9326494 Free PMC article.
-
RNA editing in plants and its evolution.Annu Rev Genet. 2013;47:335-52. doi: 10.1146/annurev-genet-111212-133519. Annu Rev Genet. 2013. PMID: 24274753 Review.
-
Evolution and mechanism of translation in chloroplasts.Annu Rev Genet. 1998;32:437-59. doi: 10.1146/annurev.genet.32.1.437. Annu Rev Genet. 1998. PMID: 9928487 Review.
Cited by
-
A ribonuclease activity linked to DYW1 in vitro is inhibited by RIP/MORF proteins.Sci Rep. 2023 Jul 3;13(1):10723. doi: 10.1038/s41598-023-36969-6. Sci Rep. 2023. PMID: 37400527 Free PMC article.
-
RNA editing site recognition in heterologous plant mitochondria.Curr Genet. 2006 Dec;50(6):405-16. doi: 10.1007/s00294-006-0100-3. Epub 2006 Oct 11. Curr Genet. 2006. PMID: 17033819
-
Formation of the Arabidopsis pentatricopeptide repeat family.Plant Physiol. 2006 Jul;141(3):825-39. doi: 10.1104/pp.106.077826. Plant Physiol. 2006. PMID: 16825340 Free PMC article.
-
Post-transcriptional control of chloroplast gene expression.Gene Regul Syst Bio. 2009 Mar 12;3:31-47. doi: 10.4137/grsb.s2080. Gene Regul Syst Bio. 2009. PMID: 19838333 Free PMC article.
-
CURE-Chloroplast: a chloroplast C-to-U RNA editing predictor for seed plants.BMC Bioinformatics. 2009 May 8;10:135. doi: 10.1186/1471-2105-10-135. BMC Bioinformatics. 2009. PMID: 19422723 Free PMC article.
References
-
- Maier, R. M., Zeltz, P., Kössel, H., Bonnard, G., Gualberto, J. M. & Grienenberger, J. M. (1996) Plant Mol. Biol. 32, 343–365. - PubMed
-
- Marchfelder, A., Binder, S., Brennicke, A. & Knoop, V. (1998) in Modification and Editing of RNA, eds. Grosjean, H. & Benne, R. (Am. Soc. Microbiol., Washington, DC), pp. 307–323.
-
- Mulligan, R. M. & Maliga, P. (1998) in A Look Beyond Transcription, eds. Bailey-Serres, J. & Gallie, D. R. (Am. Soc. Plant Physiol., Rockville, MD), pp. 153–161.
-
- Sugiura, M., Hirose, T. & Sugita, M. (1998) Annu. Rev. Genet. 32, 437–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

