Small-world communication of residues and significance for protein dynamics
- PMID: 14695252
- PMCID: PMC1303839
- DOI: 10.1016/S0006-3495(04)74086-2
Small-world communication of residues and significance for protein dynamics
Abstract
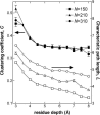
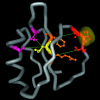
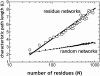
It is not merely the position of residues that is critically important for a protein's function and stability, but also their interactions. We illustrate, by using a network construction on a set of 595 nonhomologous proteins, that regular packing is preserved in short-range interactions, but short average path lengths are achieved through some long-range contacts. Thus, lying between the two extremes of regularity and randomness, residues in folded proteins are distributed according to a "small-world" topology. Using this topology, we show that the core residues have the same local packing arrangements irrespective of protein size. Furthermore, we find that the average shortest path lengths are highly correlated with residue fluctuations, providing a link between the spatial arrangement of the residues and protein dynamics.
Figures
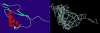



References
-
- Baase, W. A., N. C. Gassner, X.-J. Zhang, R. Kuroki, L. H. Weaver, D. E. Tronrud, and B. W. Matthews. 1999. How much sequence variation can the functions of biological molecules tolerate? In Simplicity and Complexity in Proteins and Nucleic Acid. H. Frauenfelder, J. Deisenhofer, and P. G. Wolynes, editors. Dahlem University Press, Berlin, Germany.
-
- Bagci, Z., R. L. Jernigan, and I. Bahar. 2002. Residue packing in proteins: uniform distribution on a coarse-grained scale. J. Chem. Phys. 116:2269–2276.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

