The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria
- PMID: 14699090
- PMCID: PMC2171957
- DOI: 10.1083/jcb.200310092
The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria
Abstract
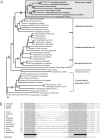
Integral proteins in the outer membrane of mitochondria control all aspects of organelle biogenesis, being required for protein import, mitochondrial fission, and, in metazoans, mitochondrial aspects of programmed cell death. How these integral proteins are assembled in the outer membrane had been unclear. In bacteria, Omp85 is an essential component of the protein insertion machinery, and we show that members of the Omp85 protein family are also found in eukaryotes ranging from plants to humans. In eukaryotes, Omp85 is present in the mitochondrial outer membrane. The gene encoding Omp85 is essential for cell viability in yeast, and conditional omp85 mutants have defects that arise from compromised insertion of integral proteins like voltage-dependent anion channel (VDAC) and components of the translocase in the outer membrane of mitochondria (TOM) complex into the mitochondrial outer membrane.
Figures

References
-
- Adams, J.M., and S. Cory. 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26:61–66. - PubMed
-
- Bay, D.C., and D.A. Court. 2002. Origami in the outer membrane: the transmembrane arrangement of mitochondrial porins. Biochem. Cell Biol. 80:551–562. - PubMed
-
- Bruno, W.J., N.D. Socci, and A.L. Halpern. 2000. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol. Biol. Evol. 17:189–197. - PubMed
-
- Buchanan, S.K. 1999. Beta-barrel proteins from bacterial outer membranes: structure, function and refolding. Curr. Opin. Struct. Biol. 9:455–461. - PubMed
-
- Cavalier-Smith, T. 2002. The phagotrophic origin of eukaryotes and phylogenetic classification of protozoa. Int. J. Syst. Evol. Microbiol. 52:297–354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

