Tap and NXT promote translation of unspliced mRNA
- PMID: 14701875
- PMCID: PMC305259
- DOI: 10.1101/gad.1155703
Tap and NXT promote translation of unspliced mRNA
Abstract
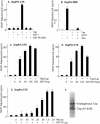
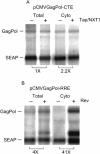
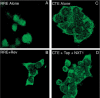
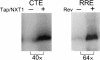
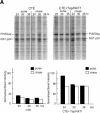
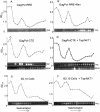
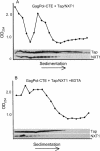
Tap has been proposed to play a role in general mRNA export and also functions in expression of RNA with retained introns that contain the MPMV CTE (constitutive transport element). Tap forms a functional heterodimer with NXT/p15. We have previously demonstrated that unspliced intron-containing CTE RNA is efficiently exported to the cytoplasm in mammalian cells. Here we show that Tap and NXT proteins function together to enhance translation of proteins from the exported CTE RNA. Pulse chase experiments show that Tap/NXT significantly increases the rate of protein synthesis. Sucrose gradient analysis demonstrates that Tap and NXT efficiently shift the unspliced RNA into polyribosomal fractions. Furthermore, Tap, but not NXT is detected in polyribosomes. Taken together, our results indicate that Tap and NXT serve a role in translational regulation of RNA after export to the cytoplasm. They further suggest that Tap/NXT may play a role in remodeling of cytoplasmic RNP complexes, providing a link between export pathways and cytoplasmic fate.
Figures

Similar articles
-
Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA.Oncogene. 2000 Jul 27;19(32):3570-5. doi: 10.1038/sj.onc.1203676. Oncogene. 2000. PMID: 10951562
-
The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element.J Biol Chem. 2007 Jul 6;282(27):19844-53. doi: 10.1074/jbc.M701660200. Epub 2007 May 18. J Biol Chem. 2007. PMID: 17513303
-
An intron with a constitutive transport element is retained in a Tap messenger RNA.Nature. 2006 Sep 14;443(7108):234-7. doi: 10.1038/nature05107. Nature. 2006. PMID: 16971948
-
Insights into the nuclear export of murine leukemia virus intron-containing RNA.RNA Biol. 2015;12(9):942-9. doi: 10.1080/15476286.2015.1065375. RNA Biol. 2015. PMID: 26158194 Free PMC article. Review.
-
Constitutive transport element-mediated nuclear export.Curr Top Microbiol Immunol. 2001;259:77-93. doi: 10.1007/978-3-642-56597-7_4. Curr Top Microbiol Immunol. 2001. PMID: 11417128 Review. No abstract available.
Cited by
-
An NXF1 mRNA with a retained intron is expressed in hippocampal and neocortical neurons and is translated into a protein that functions as an Nxf1 cofactor.Mol Biol Cell. 2016 Dec 1;27(24):3903-3912. doi: 10.1091/mbc.E16-07-0515. Epub 2016 Oct 5. Mol Biol Cell. 2016. PMID: 27708137 Free PMC article.
-
NXT2 is required for embryonic heart development in zebrafish.BMC Dev Biol. 2005 Mar 24;5:7. doi: 10.1186/1471-213X-5-7. BMC Dev Biol. 2005. PMID: 15790397 Free PMC article.
-
A novel retroviral vector system to analyze expression from mRNA with retained introns using fluorescent proteins and flow cytometry.Sci Rep. 2019 Apr 23;9(1):6467. doi: 10.1038/s41598-019-42914-3. Sci Rep. 2019. PMID: 31015546 Free PMC article.
-
The eukaryotic translation initiation factor eIF4E unexpectedly acts in splicing thereby coupling mRNA processing with translation: eIF4E induces widescale splicing reprogramming providing system-wide connectivity between splicing, nuclear mRNA export and translation.Bioessays. 2024 Jan;46(1):e2300145. doi: 10.1002/bies.202300145. Epub 2023 Nov 5. Bioessays. 2024. PMID: 37926700 Free PMC article.
-
Mining the human complexome database identifies RBM14 as an XPO1-associated protein involved in HIV-1 Rev function.J Virol. 2015 Apr;89(7):3557-67. doi: 10.1128/JVI.03232-14. Epub 2015 Jan 14. J Virol. 2015. PMID: 25589658 Free PMC article.
References
-
- Antic D. and Keene, J.D. 1998. Messenger ribonucleoprotein complexes containing human ELAV proteins: Interactions with cytoskeleton and translational apparatus. J. Cell Sci. 111 (Pt 2): 183-197. - PubMed
-
- Bachi A., Braun, I.C., Rodrigues, J.P., Pante, N., Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M., Gorlich, D., Carmo-Fonseca, M., et al. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6: 136-158. - PMC - PubMed
-
- Berkowitz R., Fisher, J., and Goff, S.P. 1996. RNA packaging. In Morphogenesis and maturation of retroviruses (ed. H.-G. Krausslich), pp. 177-218. Springer, Berlin.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
