Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling
- PMID: 14701904
- PMCID: PMC327180
- DOI: 10.1073/pnas.0304533101
Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling
Abstract
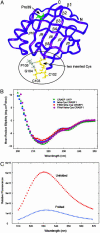
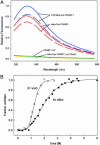
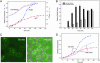
In vivo fluorescent labeling of an expressed protein has enabled the observation of its stability and aggregation directly in bacterial cells. Mammalian cellular retinoic acid-binding protein I (CRABP I) was mutated to incorporate in a surface-exposed omega loop the sequence Cys-Cys-Gly-Pro-Cys-Cys, which binds specifically to a biarsenical fluorescein dye (FlAsH). Unfolding of labeled tetra-Cys CRABP I is accompanied by enhancement of FlAsH fluorescence, which made it possible to determine the free energy of unfolding of this protein by urea titration in cells and to follow in real time the formation of inclusion bodies by a slow-folding, aggregationprone mutant (FlAsH-labeled P39A tetra-Cys CRABP I). Aggregation in vivo displayed a concentration-dependent apparent lag time similar to observations of protein aggregation in purified in vitro model systems.
Figures
References
-
- Betts, S., Haase-Pettingell, C. & King, J. A. (1997) Adv. Prot. Chem. 50, 243–264. - PubMed
-
- Fink, A. L. (1998) Folding Des. 3, R9–R23. - PubMed
-
- Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3, 906–918. - PubMed
-
- Banaszak, L., Winter, N., Xu, Z., Bernlohr, D. A., Cowan, S. & Jones, T. A. (1994) Adv. Protein Chem. 45, 89–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

