The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes
- PMID: 14701917
- PMCID: PMC316302
- DOI: 10.1104/pp.103.032904
The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes
Abstract
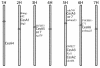
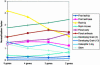
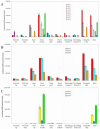
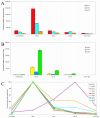
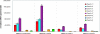
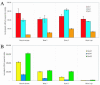
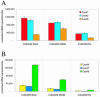
Sequence data from cDNA and genomic clones, coupled with analyses of expressed sequence tag databases, indicate that the CesA (cellulose synthase) gene family from barley (Hordeum vulgare) has at least eight members, which are distributed across the genome. Quantitative polymerase chain reaction has been used to determine the relative abundance of mRNA transcripts for individual HvCesA genes in vegetative and floral tissues, at different stages of development. To ensure accurate expression profiling, geometric averaging of multiple internal control gene transcripts has been applied for the normalization of transcript abundance. Total HvCesA mRNA levels are highest in coleoptiles, roots, and stems and much lower in floral tissues, early developing grain, and in the elongation zone of leaves. In most tissues, HvCesA1, HvCesA2, and HvCesA6 predominate, and their relative abundance is very similar; these genes appear to be coordinately transcribed. A second group, comprising HvCesA4, HvCesA7, and HvCesA8, also appears to be coordinately transcribed, most obviously in maturing stem and root tissues. The HvCesA3 expression pattern does not fall into either of these two groups, and HvCesA5 transcript levels are extremely low in all tissues. Thus, the HvCesA genes fall into two general groups of three genes with respect to mRNA abundance, and the co-expression of the groups identifies their products as candidates for the rosettes that are involved in cellulose biosynthesis at the plasma membrane. Phylogenetic analysis allows the two groups of genes to be linked with orthologous Arabidopsis CesA genes that have been implicated in primary and secondary wall synthesis.
Figures



References
-
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R et al. (1998) Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 717-720 - PubMed
-
- Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25: 169-193 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

