Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis
- PMID: 14702116
- PMCID: PMC300762
- DOI: 10.1172/JCI17803
Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis
Abstract
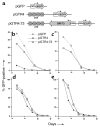
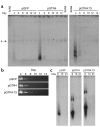
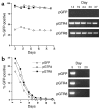
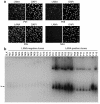
Kaposi sarcoma-associated (KS-associated) herpesvirus (KSHV) infection is linked to the development of both KS and several lymphoproliferative diseases. In all cases, the resulting tumor cells predominantly display latent viral infection. KS tumorigenesis requires ongoing lytic viral replication as well, however, for reasons that are unclear but have been suggested to involve the production of angiogenic or mitogenic factors by lytically infected cells. Here we demonstrate that proliferating cells infected with KSHV in vitro display a marked propensity to segregate latent viral genomes, with only a variable but small subpopulation being capable of stable episome maintenance. Stable maintenance is not due to the enhanced production of viral or host trans-acting factors, but is associated with cis-acting, epigenetic changes in the viral chromosome. These results indicate that acquisition of stable KSHV latency is a multistep process that proceeds with varying degrees of efficiency in different cell types. They also suggest an additional role for lytic replication in sustaining KS tumorigenesis: namely, the recruitment of new cells to latency to replace those that have segregated the viral episome.
Figures





Comment in
-
New viruses shake old paradigms.J Clin Invest. 2004 Jan;113(1):21-3. doi: 10.1172/JCI20662. J Clin Invest. 2004. PMID: 14702103 Free PMC article.
References
-
- Herndier B, Ganem D. The biology of Kaposi’s sarcoma. Cancer Treat. Res. 2001;104:89–126. - PubMed
-
- Judde JG, et al. Monoclonality or oligoclonality of human herpesvirus 8 terminal repeat sequences in Kaposi’s sarcoma and other diseases. J. Natl. Cancer Inst. 2000;92:729–736. - PubMed
-
- Corbeil J, Evans LA, Vasak E, Cooper DA, Penny R. Culture and properties of cells derived from Kaposi sarcoma. J. Immunol. 1991;146:2972–2976. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

