The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit
- PMID: 14702318
- PMCID: PMC305748
- DOI: 10.1128/JB.186.2.481-489.2004
The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit
Abstract
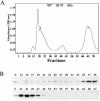
The Obg family of GTPases is widely conserved and predicted to play an as-yet-unknown role in translation. Recent reports provide circumstantial evidence that both eukaryotic and prokaryotic Obg proteins are associated with the large ribosomal subunit. Here we provide direct evidence that the Caulobacter crescentus CgtA(C) protein is associated with the free large (50S) ribosomal subunit but not with 70S monosomes or with translating ribosomes. In contrast to the Bacillus subtilis and Escherichia coli proteins, CgtA(C) does not fractionate in a large complex by gel filtration, indicating a moderately weak association with the 50S subunit. Moreover, binding of CgtA(C) to the 50S particle is sensitive to salt concentration and buffer composition but not guanine nucleotide occupancy of CgtA(C). Assays of epitope-tagged wild-type and mutant variants of CgtA(C) indicate that the C terminus of CgtA(C) is critical for 50S association. Interestingly, the addition of a C-terminal epitope tag also affected the ability of various cgtA(C) alleles to function in vivo. Depletion of CgtA(C) led to perturbations in the polysome profile, raising the possibility that CgtA(C) is involved in ribosome assembly or stability.
Figures

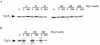
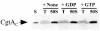




Similar articles
-
The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase.J Bacteriol. 2004 Aug;186(16):5249-57. doi: 10.1128/JB.186.16.5249-5257.2004. J Bacteriol. 2004. PMID: 15292126 Free PMC article.
-
The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels.Mol Microbiol. 2004 Dec;54(5):1379-92. doi: 10.1111/j.1365-2958.2004.04354.x. Mol Microbiol. 2004. PMID: 15554976
-
The N-terminal domain of the Caulobacter crescentus CgtA protein does not function as a guanine nucleotide exchange factor.FEBS Lett. 2000 Oct 27;484(1):29-32. doi: 10.1016/s0014-5793(00)02121-9. FEBS Lett. 2000. PMID: 11056216
-
Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis.Crit Rev Microbiol. 2014 Aug;40(3):207-24. doi: 10.3109/1040841X.2013.776510. Epub 2013 Mar 28. Crit Rev Microbiol. 2014. PMID: 23537324 Review.
-
The Obg subfamily of bacterial GTP-binding proteins: essential proteins of largely unknown functions that are evolutionarily conserved from bacteria to humans.Acta Biochim Pol. 2005;52(1):35-43. Acta Biochim Pol. 2005. PMID: 15827604 Review.
Cited by
-
Biochemical characterization of ribosome assembly GTPase RbgA in Bacillus subtilis.J Biol Chem. 2012 Mar 9;287(11):8417-23. doi: 10.1074/jbc.M111.331322. Epub 2012 Jan 20. J Biol Chem. 2012. PMID: 22267738 Free PMC article.
-
Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering.Mol Cell Proteomics. 2010 Jul;9(7):1594-615. doi: 10.1074/mcp.M000038-MCP201. Epub 2010 Apr 26. Mol Cell Proteomics. 2010. PMID: 20423899 Free PMC article.
-
Molecular modeling study for interaction between Bacillus subtilis Obg and Nucleotides.PLoS One. 2010 Sep 7;5(9):e12597. doi: 10.1371/journal.pone.0012597. PLoS One. 2010. PMID: 20830302 Free PMC article.
-
Interactions of an essential Bacillus subtilis GTPase, YsxC, with ribosomes.J Bacteriol. 2008 Jan;190(2):681-90. doi: 10.1128/JB.01193-07. Epub 2007 Nov 2. J Bacteriol. 2008. PMID: 17981968 Free PMC article.
-
Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae.Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4636-41. doi: 10.1073/pnas.0611650104. Epub 2007 Mar 5. Proc Natl Acad Sci U S A. 2007. PMID: 17360576 Free PMC article.
References
-
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. - PubMed
-
- Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP-binding protein. Structure 10:1581-1592. - PubMed
-
- Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. - PubMed
-
- Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6:135-139. - PubMed
-
- Caldon, C. E., P. Yoong, and P. E. March. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41:289-297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

