ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences
- PMID: 14704338
- PMCID: PMC373265
- DOI: 10.1093/nar/gkh152
ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences
Abstract
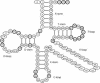
A computer program, ARAGORN, identifies tRNA and tmRNA genes. The program employs heuristic algorithms to predict tRNA secondary structure, based on homology with recognized tRNA consensus sequences and ability to form a base-paired cloverleaf. tmRNA genes are identified using a modified version of the BRUCE program. ARAGORN achieves a detection sensitivity of 99% from a set of 1290 eubacterial, eukaryotic and archaeal tRNA genes and detects all complete tmRNA sequences in the tmRNA database, improving on the performance of the BRUCE program. Recently discovered tmRNA genes in the chloroplasts of two species from the 'green' algae lineage are detected. The output of the program reports the proposed tRNA secondary structure and, for tmRNA genes, the secondary structure of the tRNA domain, the tmRNA gene sequence, the tag peptide and a list of organisms with matching tmRNA peptide tags.
Figures

References
-
- Kim S.H., Suddath,F.L., Quigley,G.J., McPherson,A., Sussman,J.L., Wang,A.H.J., Seeman,N.C. and Rich,A. (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science, 185, 435–440. - PubMed
-
- Sharp S.J., Schaak,J., Cooley,L., Burke,D.J. and Söll,D. (1985) Structure and transcription of eukaryotic tRNA genes. CRC Crit. Rev. Biochem., 19, 107–144. - PubMed
-
- Fichant G.A. and Burks,C. (1991) Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol., 220, 659–671. - PubMed
-
- Muto A., Ushida,C. and Himeno,H. (1998) A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci., 23, 25–29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

