Automatic extraction of mutations from Medline and cross-validation with OMIM
- PMID: 14704350
- PMCID: PMC373272
- DOI: 10.1093/nar/gkh162
Automatic extraction of mutations from Medline and cross-validation with OMIM
Abstract
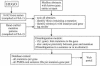
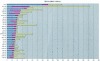
Mutations help us to understand the molecular origins of diseases. Researchers, therefore, both publish and seek disease-relevant mutations in public databases and in scientific literature, e.g. Medline. The retrieval tends to be time-consuming and incomplete. Automated screening of the literature is more efficient. We developed extraction methods (called MEMA) that scan Medline abstracts for mutations. MEMA identified 24,351 singleton mutations in conjunction with a HUGO gene name out of 16,728 abstracts. From a sample of 100 abstracts we estimated the recall for the identification of mutation-gene pairs to 35% at a precision of 93%. Recall for the mutation detection alone was >67% with a precision rate of >96%. This shows that our system produces reliable data. The subset consisting of protein sequence mutations (PSMs) from MEMA was compared to the entries in OMIM (20,503 entries versus 6699, respectively). We found 1826 PSM-gene pairs to be in common to both datasets (cross-validated). This is 27% of all PSM-gene pairs in OMIM and 91% of those pairs from OMIM which co-occur in at least one Medline abstract. We conclude that Medline covers a large portion of the mutations known to OMIM. Another large portion could be artificially produced mutations from mutagenesis experiments. Access to the database of extracted mutation-gene pairs is available through the web pages of the EBI (refer to http://www.ebi. ac.uk/rebholz/index.html).
Figures

References
-
- Perutz M.F. and Lehmann,H. (1968) Molecular pathology of human haemoglobin. Nature, 219, 902–909. - PubMed
-
- Tolle R. (2001) Information Technology Tools for Efficient SNP Studies. Am. J. Pharmacogenomics, 1, 1–12. - PubMed
-
- Medline database (December 2001) Access through US National Library of Medicine, 8600 Rockville Pike, Bethesda, MD 20894, USA.
-
- Online Mendelian Inheritance in Man, OMIM™ (December 2001) McKusick–Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD).
-
- Hamosh A., Scott,A.F., Amberger,J., Valle,D. and McKusick,V.A. (2000) Online Mendelian Inheritance in Man (OMIM). Hum. Mutat., 15, 57–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

