Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes
- PMID: 14704351
- PMCID: PMC373277
- DOI: 10.1093/nar/gkh167
Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes
Abstract
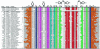
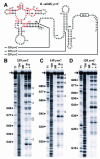
Recent studies have begun to reveal that numerous fundamental metabolic pathways in bacteria are regulated by riboswitches residing within certain messenger RNAs. These riboswitches selectively bind metabolites and modulate gene expression in response to changing ligand concentrations. Previously, we provided evidence that the btuB mRNAs of Escherichia coli and Salmonella typhimurium each carry a coenzyme B12-dependent riboswitch that causes repressed translation of the encoded cobalamin-transport protein at elevated coenzyme concentrations. Herein, we use a phylogenetic analysis to define a consensus sequence and secondary structure model for the ligand- binding domain of this riboswitch class. RNA structures that conform to this model are widespread in both Gram-positive and Gram-negative organisms. In addition, we find that the 5'-untranslated region (5'-UTR) of the cobalamin biosynthesis (cob) operon of S.typhimurium carries an RNA motif that matches this consensus sequence. Biochemical and genetic characterization of this motif confirms that the RNA directly binds coenzyme B12, and that it likely serves as a genetic control element for regulating expression of the 25-gene operon for cobalamin production in this pathogen.
Figures



References
-
- Winkler W.C. and Breaker,R.R. (2003) Genetic control by metabolite-binding riboswitches. Chembiochem, 4, 1024–1032. - PubMed
-
- Mironov A.S., Gusarov,I., Rafikov,R., Lopez,L.E., Shatalin,K., Kreneva,R.A., Perumov,D.A. and Nudler,E. (2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell, 111, 747–756. - PubMed
-
- Winkler W., Nahvi,A. and Breaker,R.R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature, 419, 952–956. - PubMed
-
- Mandal M., Boese,B., Barrick,J.E., Winkler,W.C. and Breaker,R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell, 113, 577–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

