Improved full-length cDNA production based on RNA tagging by T4 DNA ligase
- PMID: 14704363
- PMCID: PMC373303
- DOI: 10.1093/nar/gng158
Improved full-length cDNA production based on RNA tagging by T4 DNA ligase
Abstract
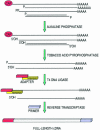
Second-strand cDNA priming is a central problem for full-length characterization of transcripts. A new strategy using bacteriophage T4 DNA ligase and partially degenerate adapters is proposed for grafting a sequence tag to the end of polyribonucleotides. Based on this RNA tagging system and previously described protocols, a new method for full-length cDNA production has been implemented. Validation of the method is shown in Arabidopsis thaliana by the construction of a full-length cDNA library and the analysis of 154 clones and by 5'-RACE-PCR run on a documented experimental system.
Figures


References
-
- Carninci P., Kvam,C., Kitamura,A., Ohsumi,T., Okazaki,Y., Itoh,M., Kamiya,M., Shibata,K., Sasaki,N., Izawa,M. et al. (1996) High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics, 37, 327–336. - PubMed
-
- Akowitz A. and Manuelidis,L. (1989) A novel cDNA/PCR strategy for efficient cloning of small amounts of undefined RNA. Gene, 81, 295–306. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

