Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors
- PMID: 14707115
- PMCID: PMC1887727
- DOI: 10.1084/jem.20031175
Gene expression profiling of hairy cell leukemia reveals a phenotype related to memory B cells with altered expression of chemokine and adhesion receptors
Abstract
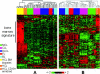
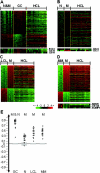
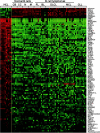
Hairy cell leukemia (HCL) is a chronic B cell malignancy characterized by the diffuse infiltration of bone marrow and spleen by cells displaying a typical "hairy" morphology. However, the nature of the HCL phenotype and its relationship to normal B cells and to other lymphoma subtypes remains unclear. Using gene expression profiling, we show here that HCL displays a homogeneous pattern of gene expression, which is clearly distinct from that of other B cell non-Hodgkin lymphomas. Comparison with the gene expression profiles of purified normal B cell subpopulations, including germinal center (GC), pre-GC (naive), and post-GC (memory) B cells, shows that HCL cells are more related to memory cells, suggesting a derivation from this B cell population. Notably, when compared with memory cells, HCL cells displayed a remarkable conservation in proliferation, apoptosis, and DNA metabolism programs, whereas they appeared significantly altered in the expression of genes controlling cell adhesion and response to chemokines. Finally, these analyses have identified several genes that are specifically expressed in HCL and whose expression was confirmed at the protein level by immunocytochemical analysis of primary HCL cases. These results have biological implications relevant to the pathogenesis of this malignancy as well as clinical implications for its diagnosis and therapy.
Figures


References
-
- Burke, J.S. 1978. The value of the bone-marrow biopsy in the diagnosis of hairy cell leukemia. Am. J. Clin. Pathol. 70:876–884. - PubMed
-
- Brunning, M., and R.W. McKenna. 1994. Atlas of Tumor Pathology: Tumors of the Bone Marrow. Armed Forces Institute of Pathology. Washington, D.C.: Universities Associated for Research and Education in Pathology.
-
- Harris, N.L., E.S. Jaffe, H. Stein, P.M. Banks, J.K. Chan, M.L. Cleary, G. Delsol, C. De Wolf-Peeters, B. Falini, and K.C. Gatter. 1994. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 84:1361–1392. - PubMed
-
- Jaffe, E.S., N.L. Harris, H. Stein, and J.W. Vardiman. 2001. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press, Lyon, France. 351 pp.
-
- Mey, U., J. Strehl, M. Gorschluter, C. Ziske, A. Glasmacher, H. Pralle, and I. Schmidt-Wolf. 2003. Advances in the treatment of hairy-cell leukaemia. Lancet. Oncol. 4:86–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

