Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish
- PMID: 14707165
- PMCID: PMC314266
- DOI: 10.1101/gr.1717804
Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish
Abstract
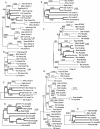
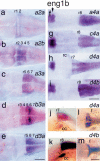
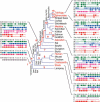
The pufferfish skeleton lacks ribs and pelvic fins, and has fused bones in the cranium and jaw. It has been hypothesized that this secondarily simplified pufferfish morphology is due to reduced complexity of the pufferfish Hox complexes. To test this hypothesis, we determined the genomic structure of Hox clusters in the Southern pufferfish Spheroides nephelus and interrogated genomic databases for the Japanese pufferfish Takifugu rubripes (fugu). Both species have at least seven Hox clusters, including two copies of Hoxb and Hoxd clusters, a single Hoxc cluster, and at least two Hoxa clusters, with a portion of a third Hoxa cluster in fugu. Results support genome duplication before divergence of zebrafish and pufferfish lineages, followed by loss of a Hoxc cluster in the pufferfish lineage and loss of a Hoxd cluster in the zebrafish lineage. Comparative analysis shows that duplicate genes continued to be lost for hundreds of millions of years, contrary to predictions for the permanent preservation of gene duplicates. Gene expression analysis in fugu embryos by in situ hybridization revealed evolutionary change in gene expression as predicted by the duplication-degeneration-complementation model. These experiments rule out the hypothesis that the simplified pufferfish body plan is due to reduction in Hox cluster complexity, and support the notion that genome duplication contributed to the radiation of teleosts into half of all vertebrate species by increasing developmental diversification of duplicate genes in daughter lineages.
Figures



References
-
- Allendorf, F. and Thorgaard, G. 1984. Tetraploidy and the evolution of salmonid fishes. In Evolutionary genetics of fishes (ed. B.J. Turner), pp. 1-46. Plenum Press, New York.
-
- Allendorf, F.W., Utter, F.M., and May, B.P. 1975. Gene duplication within the family Salmonidae: II. Detection and determination of the genetic control of duplicate loci through inheritance studies and the examination of populations. In Isozymes (ed. C.L. Markert), pp. 415-432. Academic Press, New York.
-
- Amemiya, C., Amores, A., Ota, G., Mueller, D., Garraty, J., Postlethwait, J., and Litman, G. 2001. Generation of a P1 artificial chromosome library of the Southern pufferfish. Gene 272: 283-289. - PubMed
-
- Amores, A., Force, A., Yan, Y.-L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langel, J., Prince, V., Wang, Y.-L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711-1714. - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/genome/guide/human/Q; Human Genome Resources.
-
- http://zfin.org/; The Zebrafish Information Network.
-
- http://fugu.hgmp.mrc.ac.uk/blast/; Fugu BLAST server at the UK Human Genome Mapping Project Resource Centre.
-
- http://genome.jgi-psf.org/fugu6/fugu6.home.html; Fugu rubripes at the Joint Genome Institute.
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
