An abundance of bidirectional promoters in the human genome
- PMID: 14707170
- PMCID: PMC314279
- DOI: 10.1101/gr.1982804
An abundance of bidirectional promoters in the human genome
Abstract
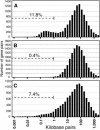
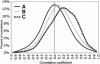
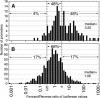
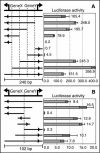
The alignment of full-length human cDNA sequences to the finished sequence of the human genome provides a unique opportunity to study the distribution of genes throughout the genome. By analyzing the distances between 23,752 genes, we identified a class of divergently transcribed gene pairs, representing more than 10% of the genes in the genome, whose transcription start sites are separated by less than 1000 base pairs. Although this bidirectional arrangement has been previously described in humans and other species, the prevalence of bidirectional gene pairs in the human genome is striking, and the mechanisms of regulation of all but a few bidirectional genes are unknown. Our work shows that the transcripts of many bidirectional pairs are coexpressed, but some are antiregulated. Further, we show that many of the promoter segments between two bidirectional genes initiate transcription in both directions and contain shared elements that regulate both genes. We also show that the bidirectional arrangement is often conserved among mouse orthologs. These findings demonstrate that a bidirectional arrangement provides a unique mechanism of regulation for a significant number of mammalian genes.
Figures
References
-
- Adachi, N. and Lieber, M.R. 2002. Bidirectional gene organization: A common architectural feature of the human genome. Cell 109: 807-809. - PubMed
-
- Ahn, J. and Gruen, J.R. 1999. The genomic organization of the histone clusters on human 6p21.3. Mamm. Genome 10: 768-770. - PubMed
-
- Albig, W., Kioschis, P., Poustka, A., Meergans, K., and Doenecke, D. 1997. Human histone gene organization: Nonregular arrangement within a large cluster. Genomics 40: 314-322. - PubMed
-
- Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.C., van Asperen, R., Boon, K., Voute, P.A., et al. 2001. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science 291: 1289-1292. - PubMed
WEB SITE REFERENCES
-
- http://www-shgc.stanford.edu/myerslab; Supplemental information.
-
- http://genome-www5.stanford.edu/MicroArray/SMD; Stanford Microarray database.
-
- http://source.stanford.edu; SOURCE.
-
- http://genome.ucsc.edu; UCSC Genome Browser.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
