Analysis of sequence variations in several human genes using phosphoramidite bond DNA fragmentation and chip-based MALDI-TOF
- PMID: 14707175
- PMCID: PMC314290
- DOI: 10.1101/gr.1653504
Analysis of sequence variations in several human genes using phosphoramidite bond DNA fragmentation and chip-based MALDI-TOF
Abstract
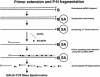
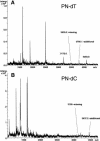
The challenge in the postgenome era is to measure sequence variations over large genomic regions in numerous patient samples. This massive amount of work can only be completed if more accurate, cost-effective, and high-throughput solutions become available. Here we describe a novel DNA fragmentation approach for single nucleotide polymorphism (SNP) discovery and sequence validation. The base-specific cleavage is achieved by creating primer extension products, in which acid-labile phosphoramidite (P-N) bonds replace the 5' phosphodiester bonds of newly incorporated pyrimidine nucleotides. Sequence variations are detected by hydrolysis of this acid-labile bond and MALDI-TOF analysis of the resulting fragments. In this study, we developed a robust protocol for P-N-bond fragmentation and investigated additional ways to improve its sensitivity and reproducibility. We also present the analysis of several human genomic targets ranging from 100-450 bp in length. By using a semiautomated sample processing protocol, we investigated an array of SNPs within a 240-bp segment of the NFKBIA gene in 48 human DNA samples. We identified and measured frequencies for the two common SNPs in the 3'UTR of NFKBIA (separated by 123 bp) and then confirmed these values in an independent genotyping experiment. The calculated allele frequencies in white and African American groups differed significantly, yet both fit Hardy-Weinberg expectations. This demonstrates the utility and effectiveness of PN-bond DNA fragmentation and subsequent MALDI-TOF MS analysis for the high-throughput discovery and measurement of sequence variations in fragments up to 0.5 kb in length in multiple human blood DNA samples.
Figures





References
-
- Agellon, L.B., Quinet, E.M., Gillette, T.G., Drayna, D.T., Brown, M.L., and Tall, A.R. 1990. Organization of the human cholesteryl ester transfer protein gene Biochemistry 29: 1372-1376. - PubMed
-
- Altshuler, D., Daly, M., and Kruglyak, L. 2000. Guilt by association. Nat. Genet. 26: 135-137. - PubMed
-
- Böcker, S. 2003. SNP and mutation discovery using base-specific cleavage and MALDI-TOF mass spectrometry. Bioinformatics 19: I44-I53. - PubMed
-
- Buetow, K.H., Edmonson, M., MacDonald, R., Clifford, R., Yip, P., Kelley, J., Little, D.P., Strausberg, R., Köster, H., Cantor, C.R., et al. 2001. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl. Acad. Sci. 98: 581-584. - PMC - PubMed
WEB SITE REFERENCES
-
- http://www.ncbi.nlm.nih.gov/SNP/; dbSNP.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
