EnsMart: a generic system for fast and flexible access to biological data
- PMID: 14707178
- PMCID: PMC314293
- DOI: 10.1101/gr.1645104
EnsMart: a generic system for fast and flexible access to biological data
Abstract
The EnsMart system (www.ensembl.org/EnsMart) provides a generic data warehousing solution for fast and flexible querying of large biological data sets and integration with third-party data and tools. The system consists of a query-optimized database and interactive, user-friendly interfaces. EnsMart has been applied to Ensembl, where it extends its genomic browser capabilities, facilitating rapid retrieval of customized data sets. A wide variety of complex queries, on various types of annotations, for numerous species are supported. These can be applied to many research problems, ranging from SNP selection for candidate gene screening, through cross-species evolutionary comparisons, to microarray annotation. Users can group and refine biological data according to many criteria, including cross-species analyses, disease links, sequence variations, and expression patterns. Both tabulated list data and biological sequence output can be generated dynamically, in HTML, text, Microsoft Excel, and compressed formats. A wide range of sequence types, such as cDNA, peptides, coding regions, UTRs, and exons, with additional upstream and downstream regions, can be retrieved. The EnsMart database can be accessed via a public Web site, or through a Java application suite. Both implementations and the database are freely available for local installation, and can be extended or adapted to 'non-Ensembl' data sets.
Figures
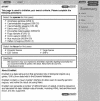



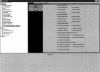


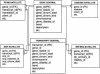
References
-
- Devlin, B. 1997. Data warehouse. From architecture to implementation, chapter 2. Addison Wesley Longman, Inc., Reading, MA.
WEB SITE REFERENCES
-
- www.ebi.ac.uk/miamexpress; MIAMExpress.
-
- www.rzpd.de/colBox/html/; RZPD's Genome-Matrix.
-
- www.ncbi.nlm.nih.gov; MapViewer at NCBI.
-
- www.ensembl.org/EnsMart; EnsMart.
-
- www.sanger.ac.uk; The Vertebrate Genome Annotation database.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
