Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae
- PMID: 14709174
- PMCID: PMC395734
- DOI: 10.1186/gb-2003-5-1-r2
Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae
Abstract
Background: Although the protein-coding sequences in the Saccharomyces cerevisiae genome have been studied and annotated extensively, much less is known about the extent and characteristics of the untranslated regions of yeast mRNAs.
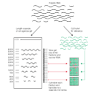
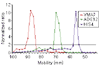
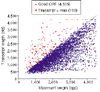
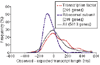
Results: We developed a 'Virtual Northern' method, using DNA microarrays for genome-wide systematic analysis of mRNA lengths. We used this method to measure mRNAs corresponding to 84% of the annotated open reading frames (ORFs) in the S. cerevisiae genome, with high precision and accuracy (measurement errors +/- 6-7%). We found a close linear relationship between mRNA lengths and the lengths of known or predicted translated sequences; mRNAs were typically around 300 nucleotides longer than the translated sequences. Analysis of genes deviating from that relationship identified ORFs with annotation errors, ORFs that appear not to be bona fide genes, and potentially novel genes. Interestingly, we found that systematic differences in the total length of the untranslated sequences in mRNAs were related to the functions of the encoded proteins.
Conclusions: The Virtual Northern method provides a practical and efficient method for genome-scale analysis of transcript lengths. Approximately 12-15% of the yeast genome is represented in untranslated sequences of mRNAs. A systematic relationship between the lengths of the untranslated regions in yeast mRNAs and the functions of the proteins they encode may point to an important regulatory role for these sequences.
Figures



Similar articles
-
Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis.Yeast. 1999 Feb;15(3):219-53. doi: 10.1002/(SICI)1097-0061(199902)15:3<219::AID-YEA349>3.0.CO;2-3. Yeast. 1999. PMID: 10077188
-
Transcript map of two regions from chromosome XI of Saccharomyces cerevisiae for interpretation of systematic sequencing results.Yeast. 1994 Nov;10(11):1403-13. doi: 10.1002/yea.320101103. Yeast. 1994. PMID: 7871880
-
Virtual Northern analysis of the human genome.PLoS One. 2007 May 23;2(5):e460. doi: 10.1371/journal.pone.0000460. PLoS One. 2007. PMID: 17520019 Free PMC article.
-
Life with 6000 genes.Science. 1996 Oct 25;274(5287):546, 563-7. doi: 10.1126/science.274.5287.546. Science. 1996. PMID: 8849441 Review.
-
Sequence organization of the mitochondrial genome of yeast--a review.Gene. 1985;37(1-3):1-17. doi: 10.1016/0378-1119(85)90252-5. Gene. 1985. PMID: 3902568 Review.
Cited by
-
Weak 5'-mRNA secondary structures in short eukaryotic genes.Genome Biol Evol. 2012;4(10):1046-53. doi: 10.1093/gbe/evs082. Genome Biol Evol. 2012. PMID: 23034215 Free PMC article.
-
Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae.Nucleic Acids Res. 2008 Jan;36(1):144-56. doi: 10.1093/nar/gkm986. Epub 2007 Nov 13. Nucleic Acids Res. 2008. PMID: 17999996 Free PMC article.
-
Multiple transcripts from a 3'-UTR reporter vary in sensitivity to nonsense-mediated mRNA decay in Saccharomyces cerevisiae.PLoS One. 2013 Nov 18;8(11):e80981. doi: 10.1371/journal.pone.0080981. eCollection 2013. PLoS One. 2013. PMID: 24260526 Free PMC article.
-
Comparative analysis of structured RNAs in S. cerevisiae indicates a multitude of different functions.BMC Biol. 2007 Jun 18;5:25. doi: 10.1186/1741-7007-5-25. BMC Biol. 2007. PMID: 17577407 Free PMC article.
-
Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide.Mol Cell Biol. 2005 Nov;25(21):9447-59. doi: 10.1128/MCB.25.21.9447-9459.2005. Mol Cell Biol. 2005. PMID: 16227595 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

