Oriented clonal cell growth in the developing mouse myocardium underlies cardiac morphogenesis
- PMID: 14709543
- PMCID: PMC2171975
- DOI: 10.1083/jcb.200309160
Oriented clonal cell growth in the developing mouse myocardium underlies cardiac morphogenesis
Abstract
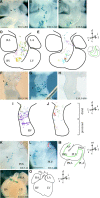
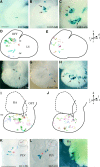
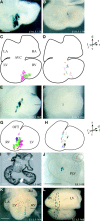
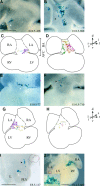
During heart morphogenesis, cardiac chambers arise by differential expansion of regions of the primitive cardiac tube. This process is under the control of specific transcription factors such as Tbx5 and dHAND. To gain insight into the cellular mechanisms that underlie cardiogenesis, we have used a retrospective clonal approach based on the spontaneous recombination of an nlaacZ reporter gene targeted to the murine alpha-cardiac actin locus. We show that clonal growth of myocardial cells is oriented. At embryonic day (E) 10.5, the shape of clones is characteristic of a given cardiac region and reflects its morphology. This is already detectable in the primitive cardiac tube at E8.5, and is maintained after septation at E14.5 with additional modulations. The clonal analysis reveals new subdivisions of the myocardium, including an interventricular boundary region. Our results show that the myocardium, from the time of its formation, is a polarized and regionalized tissue and point to the role of oriented clonal cell growth in cardiac chamber morphogenesis.
Figures



References
-
- Bonnerot, C., and J.F. Nicolas. 1993. Clonal analysis in the intact mouse embryo by intragenic homologous recombination. C. R. Acad. Sci. III. 316:1207–1217. - PubMed
-
- Bruneau, B., M. Logan, N. Davis, T. Levi, C. Tabin, J. Seidman, and C. Seidman. 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 211:100–108. - PubMed
-
- Campione, M., M.A. Ros, J.M. Icardo, E. Piedra, V.M. Christoffels, A. Schweickert, M. Blum, D. Franco, and A.F. Moorman. 2001. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev. Biol. 231:252–264. - PubMed
-
- Chen, F., H. Kook, R. Milewski, A. Gitler, M. Lu, J. Li, R. Nazarian, R. Schnepp, K. Jen, C. Biben, et al. 2002. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 110:713–723. - PubMed
-
- Chen, T.H., T.C. Chang, J.O. Kang, B. Choudhary, T. Makita, C.M. Tran, J.B. Burch, H. Eid, and H.M. Sucov. 2002. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev. Biol. 250:198–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

