Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays
- PMID: 14709637
- PMCID: PMC7108136
- DOI: 10.1373/clinchem.2003.023663
Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays
Abstract
Background: A novel coronavirus (CoV) was recently identified as the agent for severe acute respiratory syndrome (SARS). We compared the abilities of conventional and real-time reverse transcription-PCR (RT-PCR) assays to detect SARS CoV in clinical specimens.
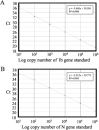
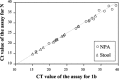
Methods: RNA samples isolated from nasopharyngeal aspirate (NPA; n = 170) and stool (n = 44) were reverse-transcribed and tested by our in-house conventional RT-PCR assay. We selected 98 NPA and 37 stool samples collected at different times after the onset of disease and tested them in a real-time quantitative RT-PCR specific for the open reading frame (ORF) 1b region of SARS CoV. Detection rates for the conventional and real-time quantitative RT-PCR assays were compared. To investigate the nature of viral RNA molecules in these clinical samples, we determined copy numbers of ORF 1b and nucleocapsid (N) gene sequences of SARS CoV.
Results: The quantitative real-time RT-PCR assay was more sensitive than the conventional RT-PCR assay for detecting SARS CoV in samples collected early in the course of the disease. Real-time assays targeted at the ORF 1b region and the N gene revealed that copy numbers of ORF 1b and N gene sequences in clinical samples were similar.
Conclusions: NPA and stool samples can be used for early diagnosis of SARS. The real-time quantitative RT-PCR assay for SARS CoV is potentially useful for early detection of SARS CoV. Our results suggest that genomic RNA is the predominant viral RNA species in clinical samples.
Figures

References
-
- World Health Organization. Acute respiratory syndrome. Wkly Epidemiol Rec 2003;78:73-74. - PubMed
-
- World Health Organization. Communicable disease surveillance & response (CSR). Cumulative number of reported probable cases. http://www.who.int/csr/sars/country/2003_06_17/en/ (Accessed June 18, 2003)..
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-1966. - PubMed
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348:1967-1976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

