Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution
- PMID: 14710190
- PMCID: PMC1298953
- DOI: 10.1038/sj.embor.7400036
Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution
Abstract
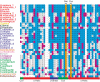
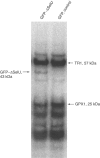
While the genome sequence and gene content are available for an increasing number of organisms, eukaryotic selenoproteins remain poorly characterized. The dual role of the UGA codon confounds the identification of novel selenoprotein genes. Here, we describe a comparative genomics approach that relies on the genome-wide prediction of genes with in-frame TGA codons, and the subsequent comparison of predictions from different genomes, wherein conservation in regions flanking the TGA codon suggests selenocysteine coding function. Application of this method to human and fugu genomes identified a novel selenoprotein family, named SelU, in the puffer fish. The selenocysteine-containing form also occurred in other fish, chicken, sea urchin, green algae and diatoms. In contrast, mammals, worms and land plants contained cysteine homologues. We demonstrated selenium incorporation into chicken SelU and characterized the SelU expression pattern in zebrafish embryos. Our data indicate a scattered evolutionary distribution of selenoproteins in eukaryotes, and suggest that, contrary to the picture emerging from data available so far, other taxa-specific selenoproteins probably exist.
Figures



References
-
- Abril JF, Guigó R (2000) gff2ps: visualizing genomic annotations. Bioinformatics 16: 743–744 - PubMed
-
- Berry MJ, Mai AL, Kieffer J, Harney JW, Larsen P (1992) Substitution of cysteine for selenocysteine in type i iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology 131: 1848–1852 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

