Selection and characterization of conditionally active promoters in Lactobacillus plantarum, using alanine racemase as a promoter probe
- PMID: 14711657
- PMCID: PMC321294
- DOI: 10.1128/AEM.70.1.310-317.2004
Selection and characterization of conditionally active promoters in Lactobacillus plantarum, using alanine racemase as a promoter probe
Abstract
This paper describes the use of the alr gene, encoding alanine racemase, as a promoter-screening tool for the identification of conditional promoters in Lactobacillus plantarum. Random fragments of the L. plantarum WCFS1 genome were cloned upstream of the promoterless alr gene of Lactococcus lactis in a low-copy-number plasmid vector. The resulting plasmid library was introduced into an L. plantarum Deltaalr strain (MD007), and 40,000 clones were selected. The genome coverage of the library was estimated to be 98%, based on nucleotide insert sequence and restriction analyses of the inserts of randomly selected clones. The library was screened for clones that were capable of complementing the D-alanine auxotroph phenotype of MD007 in media containing up to 10, 100, or 300 micro g of the competitive Alr inhibitor D-cycloserine per ml. Western blot analysis with polyclonal antibodies raised against lactococcal Alr revealed that the Alr production level required for growth increased in the presence of increasing concentrations of D-cycloserine, adding a quantitative factor to the primarily qualitative nature of the alr complementation screen. Screening of the alr complementation library for clones that could grow only in the presence of 0.8 M NaCl resulted in the identification of eight clones that upon Western blot analysis showed significantly higher Alr production under high-salt conditions than under low-salt conditions. These results established the effectiveness of the alanine racemase complementation screening method for the identification of promoters on their conditional or constitutive activity.
Figures
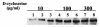
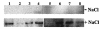
References
-
- Achen, M. G., B. E. Davidson, and A. J. Hillier. 1986. Construction of plasmid vectors for the detection of streptococcal promoters. Gene 45:45-49. - PubMed
-
- Bailey, T. L., M. E. Baker, and C. P. Elkan. 1997. An artificial intelligence approach to motif discovery in protein sequences: application to steriod dehydrogenases. J. Steroid Biochem. Mol. Biol. 62:29-44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

