PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450
- PMID: 14711998
- PMCID: PMC321774
- DOI: 10.1073/pnas.2636936100
PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450
Abstract
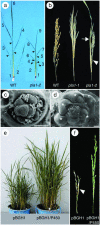
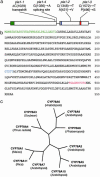
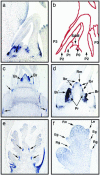
During postembryonic development of higher plants, the shoot apical meristem produces lateral organs in a regular spacing (phyllotaxy) and a regular timing (plastochron). Molecular analysis of mutants associated with phyllotaxy and plastochron would greatly increase understanding of the developmental mechanism of plant architecture because phyllotaxy and plastochron are fundamental regulators of plant architecture. pla1 of rice is not only a plastochron mutant showing rapid leaf initiation without affecting phyllotaxy, but also a heterochronic mutant showing ectopic shoot formation in the reproductive phase. Thus, pla1 provides a tool for analyzing the molecular basis of temporal regulation in leaf development. In this work, we isolated the PLA1 gene by map-based cloning. The identified PLA1 gene encodes a cytochrome P450, CYP78A11, which potentially catalyzes substances controlling plant development. PLA1 is expressed in developing leaf primordia, bracts of the panicle, and elongating internodes, but not in the shoot apical meristem. The expression pattern and mutant phenotype suggest that the PLA1 gene acting in developing leaf primordia affects the timing of successive leaf initiation and the termination of vegetative growth.
Figures
Similar articles
-
Compact shoot and leafy head 1, a mutation affects leaf initiation and developmental transition in rice (Oryza sativa L).Plant Cell Rep. 2007 Apr;26(4):421-7. doi: 10.1007/s00299-006-0259-6. Epub 2006 Nov 17. Plant Cell Rep. 2007. PMID: 17111113
-
PLASTOCHRON2 regulates leaf initiation and maturation in rice.Plant Cell. 2006 Mar;18(3):612-25. doi: 10.1105/tpc.105.037622. Epub 2006 Feb 3. Plant Cell. 2006. PMID: 16461585 Free PMC article.
-
Regulation of the plastochron by three many-noded dwarf genes in barley.PLoS Genet. 2021 May 10;17(5):e1009292. doi: 10.1371/journal.pgen.1009292. eCollection 2021 May. PLoS Genet. 2021. PMID: 33970916 Free PMC article.
-
Genetic and molecular analysis of patterning in plant development.ASGSB Bull. 1995 Oct;8(2):29-37. ASGSB Bull. 1995. PMID: 11538548 Review.
-
Formation of primordia and phyllotaxy.Curr Opin Plant Biol. 2005 Feb;8(1):53-8. doi: 10.1016/j.pbi.2004.11.013. Curr Opin Plant Biol. 2005. PMID: 15653400 Review.
Cited by
-
Over-expression of microRNA171 affects phase transitions and floral meristem determinancy in barley.BMC Plant Biol. 2013 Jan 7;13:6. doi: 10.1186/1471-2229-13-6. BMC Plant Biol. 2013. PMID: 23294862 Free PMC article.
-
Establishment of the Embryonic Shoot Meristem Involves Activation of Two Classes of Genes with Opposing Functions for Meristem Activities.Int J Mol Sci. 2020 Aug 15;21(16):5864. doi: 10.3390/ijms21165864. Int J Mol Sci. 2020. PMID: 32824181 Free PMC article.
-
Comparative transcriptome analysis reveals candidate genes for cold stress response and early flowering in pineapple.Sci Rep. 2023 Nov 2;13(1):18890. doi: 10.1038/s41598-023-45722-y. Sci Rep. 2023. PMID: 37919298 Free PMC article.
-
Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean.Plant Mol Biol. 2016 Jan;90(1-2):33-47. doi: 10.1007/s11103-015-0392-0. Epub 2015 Oct 19. Plant Mol Biol. 2016. PMID: 26482479
-
The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis.Plant Mol Biol. 2008 May;67(1-2):183-95. doi: 10.1007/s11103-008-9310-z. Epub 2008 Feb 17. Plant Mol Biol. 2008. PMID: 18278578 Free PMC article.
References
-
- Walbot, V. (1985) Trends Genet. 1, 165–169.
-
- Steeves, T. A. & Sussex, I. M. (1989) in Patterns in Plant Development (Cambridge Univ. Press, Cambridge, U.K.), pp. 46–310.
-
- Sussex, I. M. (1989) Cell 56, 225–229. - PubMed
-
- Lyndon, R. F. (1998) in The Shoot Apical Meristem: Its Growth and Development (Cambridge Univ. Press, Cambridge, U.K.), pp. 142–193.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

