Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2
- PMID: 14713949
- PMCID: PMC1271668
- DOI: 10.1038/sj.emboj.7600030
Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2
Abstract
Transient phosphorylation of the alpha-subunit of translation initiation factor 2 (eIF2alpha) represses translation and activates select gene expression under diverse stressful conditions. Defects in the eIF2alpha phosphorylation-dependent integrated stress response impair resistance to accumulation of malfolded proteins in the endoplasmic reticulum (ER stress), to oxidative stress and to nutrient deprivations. To study the hypothesized protective role of eIF2alpha phosphorylation in isolation of parallel stress signaling pathways, we fused the kinase domain of pancreatic endoplasmic reticulum kinase (PERK), an ER stress-inducible eIF2alpha kinase that is normally activated by dimerization, to a protein module that binds a small dimerizer molecule. The activity of this artificial eIF2alpha kinase, Fv2E-PERK, is subordinate to the dimerizer and is uncoupled from upstream stress signaling. Fv2E-PERK activation enhanced the expression of numerous stress-induced genes and protected cells from the lethal effects of oxidants, peroxynitrite donors and ER stress. Our findings indicate that eIF2alpha phosphorylation can initiate signaling in a cytoprotective gene expression pathway independently of other parallel stress-induced signals and that activation of this pathway can single-handedly promote a stress-resistant preconditioned state.
Figures
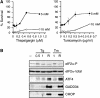


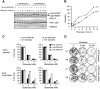
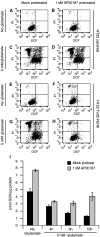

References
-
- Anderson ME 1985. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol 113: 548–555 - PubMed
-
- Bertolotti A, Zhang Y, Hendershot L, Harding H, Ron D 2000. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat Cell Biol 2: 326–332 - PubMed
-
- Brostrom CO, Brostrom MA 1998. Regulation of translational initiation during cellular responses to stress. Prog Nucleic Acid Res Mol Biol 58: 79–125 - PubMed
-
- Chen J 2000. Heme-regulated eIF2α kinase. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, and Mathews MB (eds) pp 529–546. Cold Spring Harbor: CSHL Press
-
- Davis JB, Maher P 1994. Protein kinase C activation inhibits glutamate-induced cytotoxicity in a neuronal cell line. Brain Res 652: 169–173 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

