Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource
- PMID: 14715089
- PMCID: PMC331396
- DOI: 10.1186/1471-2105-5-4
Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource
Abstract
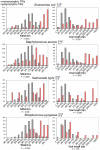
Background: Polymorphic tandem repeat typing is a new generic technology which has been proved to be very efficient for bacterial pathogens such as B. anthracis, M. tuberculosis, P. aeruginosa, L. pneumophila, Y. pestis. The previously developed tandem repeats database takes advantage of the release of genome sequence data for a growing number of bacteria to facilitate the identification of tandem repeats. The development of an assay then requires the evaluation of tandem repeat polymorphism on well-selected sets of isolates. In the case of major human pathogens, such as S. aureus, more than one strain is being sequenced, so that tandem repeats most likely to be polymorphic can now be selected in silico based on genome sequence comparison.
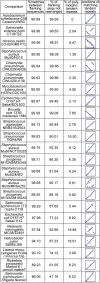
Results: In addition to the previously described general Tandem Repeats Database, we have developed a tool to automatically identify tandem repeats of a different length in the genome sequence of two (or more) closely related bacterial strains. Genome comparisons are pre-computed. The results of the comparisons are parsed in a database, which can be conveniently queried over the internet according to criteria of practical value, including repeat unit length, predicted size difference, etc. Comparisons are available for 16 bacterial species, and the orthopox viruses, including the variola virus and three of its close neighbors.
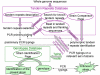
Conclusions: We are presenting an internet-based resource to help develop and perform tandem repeats based bacterial strain typing. The tools accessible at http://minisatellites.u-psud.fr now comprise four parts. The Tandem Repeats Database enables the identification of tandem repeats across entire genomes. The Strain Comparison Page identifies tandem repeats differing between different genome sequences from the same species. The "Blast in the Tandem Repeats Database" facilitates the search for a known tandem repeat and the prediction of amplification product sizes. The "Bacterial Genotyping Page" is a service for strain identification at the subspecies level.
Figures



References
-
- Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, Cousins DV, Graviss EA, Shashkina E, Kreiswirth BN, Musser JM. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics. 2002;162:1533–43. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

