Using 3D Hidden Markov Models that explicitly represent spatial coordinates to model and compare protein structures
- PMID: 14715091
- PMCID: PMC344530
- DOI: 10.1186/1471-2105-5-2
Using 3D Hidden Markov Models that explicitly represent spatial coordinates to model and compare protein structures
Abstract
Background: Hidden Markov Models (HMMs) have proven very useful in computational biology for such applications as sequence pattern matching, gene-finding, and structure prediction. Thus far, however, they have been confined to representing 1D sequence (or the aspects of structure that could be represented by character strings).
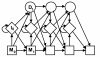
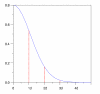
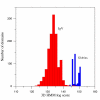
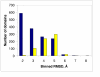
Results: We develop an HMM formalism that explicitly uses 3D coordinates in its match states. The match states are modeled by 3D Gaussian distributions centered on the mean coordinate position of each alpha carbon in a large structural alignment. The transition probabilities depend on the spread of the neighboring match states and on the number of gaps found in the structural alignment. We also develop methods for aligning query structures against 3D HMMs and scoring the result probabilistically. For 1D HMMs these tasks are accomplished by the Viterbi and forward algorithms. However, these will not work in unmodified form for the 3D problem, due to non-local quality of structural alignment, so we develop extensions of these algorithms for the 3D case. Several applications of 3D HMMs for protein structure classification are reported. A good separation of scores for different fold families suggests that the described construct is quite useful for protein structure analysis.
Conclusion: We have created a rigorous 3D HMM representation for protein structures and implemented a complete set of routines for building 3D HMMs in C and Perl. The code is freely available from http://www.molmovdb.org/geometry/3dHMM, and at this site we also have a simple prototype server to demonstrate the features of the described approach.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

