PCR-based genotyping of Mycobacterium tuberculosis with new GC-rich repeated sequences and IS6110 inverted repeats used as primers
- PMID: 14715782
- PMCID: PMC321654
- DOI: 10.1128/JCM.42.1.372-377.2004
PCR-based genotyping of Mycobacterium tuberculosis with new GC-rich repeated sequences and IS6110 inverted repeats used as primers
Abstract
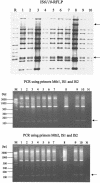
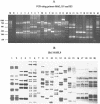
In the present study we attempted to develop a PCR-based epidemiological tool for the differentiation of Mycobacterium tuberculosis isolates. Use of the designed primers Mtb1 (5'-CCG-GCG-GGG-CCG-GCG-G) and Mtb2 (5'-CGG-CGG-CAA-CGG-CGG-C) targeting frequently repeated 16-bp sequences in combination with primers sited at the inverted repeats flanking IS6110 allowed differentiation of M. tuberculosis isolates.
Figures



References
-
- Athwal, R. S., S. S. Deo, and T. Imaeda. 1984. Deoxyribonucleic acid relatedness among Mycobacterium leprae, Mycobacterium lepraemurium and selected bacteria by dot blot and spectrophotometric deoxyribonucleic acid hybridization assays. Int. J. Syst. Bacteriol. 34:371-375.
-
- Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 11:537-544. - PubMed
-
- Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. - PubMed
-
- du Plessis, D. G., R. Warren, M. Richardson, J. J. Joubert, and P. D. van Helden. 2001. Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS6110 and other repetitive element-based DNA fingerprinting. Tuberculosis 81:211-220. - PubMed
-
- Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

