Development of a single-tube, cell lysis-based, genus-specific PCR method for rapid identification of mycobacteria: optimization of cell lysis, PCR primers and conditions, and restriction pattern analysis
- PMID: 14715804
- PMCID: PMC321669
- DOI: 10.1128/JCM.42.1.453-457.2004
Development of a single-tube, cell lysis-based, genus-specific PCR method for rapid identification of mycobacteria: optimization of cell lysis, PCR primers and conditions, and restriction pattern analysis
Abstract
A single-tube PCR method was developed for efficient identification of nontuberculous mycobacteria (NTM) and their environmental isolates in about 3 h without conventional DNA isolation. The following three steps were optimized or developed: (i). a simple, 6-min direct cell lysis protocol as a PCR prestep for generation of DNA-template, (ii). an improved Mycobacterium-specific PCR amplification protocol with a broader species specificity using newly designed primers targeting a 228-bp region of the 65-kDa heat shock protein (hsp) gene and optimal PCR amplification conditions, and (iii). a genus-specific restriction analysis of the PCR product for conclusive identification of the unknown NTM isolates.
Figures
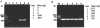


References
-
- Armand, M. O., M. Mestdagh, and F. Porteals. 2001. DNA isolation from chloroform/methanol-treated mycobacterial cells without lysozyme and proteinase K. BioTechniques 30:272-274. - PubMed
-
- Dailloux, M., C. Laurain, M. Weber, and P. Hartemann. 1999. Water and nontuberculosis mycobacteria. Water Res. 33:2219-2228.
-
- De Baere, T., R. de Mendonca, G. Claeys, G. Verschraegen, W. Mijs, R. Verhelst, S. Rottiers, L. Van Simaey, C. De Ganck, and M. Vaneechoutte. 2002. Evaluation of amplified rDNA restriction analysis (ARDRA) for the identification of cultured mycobacteria in a diagnostic laboratory. BMC Microbiol. 2:4-15. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

