The ABRF-MIRG'02 study: assembly state, thermodynamic, and kinetic analysis of an enzyme/inhibitor interaction
- PMID: 14715884
- PMCID: PMC2279960
The ABRF-MIRG'02 study: assembly state, thermodynamic, and kinetic analysis of an enzyme/inhibitor interaction
Abstract
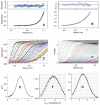
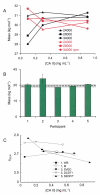
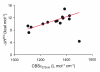
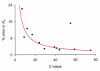
Fully characterizing the interactions involving biomolecules requires information on the assembly state, affinity, kinetics, and thermodynamics associated with complex formation. The analytical technologies often used to measure biomolecular interactions include analytical ultracentrifugation (AUC), isothermal titration calorimetry (ITC), and surface plasmon resonance (SPR). In order to evaluate the capabilities of core facilities to implement these technologies, the Association of Biomolecular Resource Facilities (ABRF) Molecular Interactions Research Group (MIRG) developed a standardized model system and distributed it to a panel of AUC, ITC, and SPR operators. The model system was composed of a well-characterized enzyme-inhibitor pair, namely bovine carbonic anhydrase II (CA II) and 4-carboxybenzenesulfonamide (CBS). Study participants were asked to measure one or more of the following: (1) the molecular mass, homogeneity, and assembly state of CA II by AUC; (2) the affinity and thermodynamics for complex formation by ITC; and (3) the affinity and kinetics of complex formation by SPR. The results from this study provide a benchmark for comparing the capabilities of individual laboratories and for defining the utility of the different instrumentation.
Figures



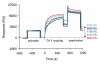
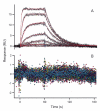

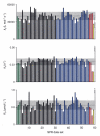
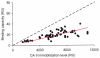

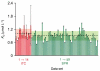
References
-
- Perkins SJ. X-ray and neutron scattering analyses of hydration shells: A molecular interpretation based on sequence predictions and modelling fits. Biophys Chem 2001;93:129–139. - PubMed
-
- Stafford WF 3rd. Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem 1992;203:295–301. - PubMed
-
- Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem 2000;279:151–163. - PubMed
-
- Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers 2000;54:328–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
