The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety
- PMID: 14715897
- PMCID: PMC321776
- DOI: 10.1073/pnas.0304808101
The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety
Abstract
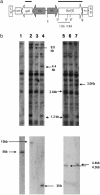
The Vf gene from the wild species Malus floribunda 821 is the most studied apple scab resistance gene. Several molecular markers mapping around this gene were the starting point for a positional cloning project. The analysis of the bacterial artificial chromosome clones spanning the Vf region led to the identification of a cluster of genes homologous to the Cladosporium fulvum resistance gene family of tomato. One of these genes, HcrVf2 (homologue of the C. fulvum resistance genes of the Vf region), was used to transform the susceptible apple cultivar Gala. Four independent transformed lines resistant to apple scab were produced, proving that HcrVf2 is sufficient to confer scab resistance to a susceptible cultivar. The results show that direct gene transfer between cross-compatible species can be viable when, as in apple, the use of backcrosses to introduce resistance genes from wild species cannot exactly reconstitute the heterozygous genotype of clonally propagated cultivars.
Figures

References
-
- Williams, E. B. & Kuc, J. (1969) Annu. Rev. Phytopathol. 7, 223–246.
-
- Janick, J., Cummins, J. N., Brown, S. K. & Hemmat, M. (1996) Fruit Breeding: Tree and Tropical Fruits (Wiley, New York), pp. 1–77.
-
- Parisi, L., Lespinasse, Y., Guillaumes, J. & Kruger, J. (1993) Phytopathology 83, 533–537.
-
- Bénaouf, G. & Parisi, L. (2000) Phytopathology 90, 236–242. - PubMed
-
- MacHardy, W. E., Gadoury, D. M. & Gessler, C. (2001) Plant Dis. 85, 1036–1051. - PubMed

