Decoding the genome: a modified view
- PMID: 14715921
- PMCID: PMC384350
- DOI: 10.1093/nar/gkh185
Decoding the genome: a modified view
Abstract
Transfer RNA's role in decoding the genome is critical to the accuracy and efficiency of protein synthesis. Though modified nucleosides were identified in RNA 50 years ago, only recently has their importance to tRNA's ability to decode cognate and wobble codons become apparent. RNA modifications are ubiquitous. To date, some 100 different posttranslational modifications have been identified. Modifications of tRNA are the most extensively investigated; however, many other RNAs have modified nucleosides. The modifications that occur at the first, or wobble position, of tRNA's anticodon and those 3'-adjacent to the anticodon are of particular interest. The tRNAs most affected by individual and combinations of modifications respond to codons in mixed codon boxes where distinction of the third codon base is important for discriminating between the correct cognate or wobble codons and the incorrect near-cognate codons (e.g. AAA/G for lysine versus AAU/C asparagine). In contrast, other modifications expand wobble codon recognition, such as U*U base pairing, for tRNAs that respond to multiple codons of a 4-fold degenerate codon box (e.g. GUU/A/C/G for valine). Whether restricting codon recognition, expanding wobble, enabling translocation, or maintaining the messenger RNA, reading frame modifications appear to reduce anticodon loop dynamics to that accepted by the ribosome. Therefore, we suggest that anticodon stem and loop domain nucleoside modifications allow a limited number of tRNAs to accurately and efficiently decode the 61 amino acid codons by selectively restricting some anticodon-codon interactions and expanding others.
Figures

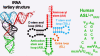

References
-
- Crick F.H.C. (1957) In The Structure of Nucleic Acids and their Role in Protein Synthesis. Biochem. Soc. Symp. 14. Cambridge University Press, UK, pp. 25–26. - PubMed
-
- Crick F.H.C., Barnett,L., Brenner,S. and Watts-Tobin,R.J. (1961) General nature of the genetic code for proteins. Nature, 192, 1227–1232. - PubMed
-
- Streisinger G., Okada,Y., Emrich,J., Newton,J., Tsugita,A., Terzaghi,E. and Inouye,M. (1966) Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol., 31, 77–84. - PubMed
-
- Nirenberg M., Caskey,T., Marshall,R., Brimacombe,R., Kellogg,D., Doctor,B., Hatfield,D., Levin,J., Rottman,F., Pestka,S., Wilcox,M. and Anderson,F. (1966) The RNA code and protein synthesis (1966) Cold Spring Harbor Symp. Quant. Biol., 31, 11–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

