The single-stranded DNA-binding protein of Deinococcus radiodurans
- PMID: 14718065
- PMCID: PMC331404
- DOI: 10.1186/1471-2180-4-2
The single-stranded DNA-binding protein of Deinococcus radiodurans
Abstract
Background: Deinococcus radiodurans R1 is one of the most radiation-resistant organisms known and is able to repair an unusually large amount of DNA damage without induced mutation. Single-stranded DNA-binding (SSB) protein is an essential protein in all organisms and is involved in DNA replication, recombination and repair. The published genomic sequence from Deinococcus radiodurans includes a putative single-stranded DNA-binding protein gene (ssb; DR0100) requiring a translational frameshift for synthesis of a complete SSB protein. The apparently tripartite gene has inspired considerable speculation in the literature about potentially novel frameshifting or RNA editing mechanisms. Immediately upstream of the ssb gene is another gene (DR0099) given an ssb-like annotation, but left unexplored.
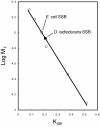
Results: A segment of the Deinococcus radiodurans strain R1 genome encompassing the ssb gene has been re-sequenced, and two errors involving omitted guanine nucleotides have been documented. The corrected sequence incorporates both of the open reading frames designated DR0099 and DR0100 into one contiguous ssb open reading frame (ORF). The corrected gene requires no translational frameshifts and contains two predicted oligonucleotide/oligosaccharide-binding (OB) folds. The protein has been purified and its sequence is closely related to the Thermus thermophilus and Thermus aquaticus SSB proteins. Like the Thermus SSB proteins, the SSBDr functions as a homodimer. The Deinococcus radiodurans SSB homodimer stimulates Deinococcus radiodurans RecA protein and Escherichia coli RecA protein-promoted DNA three-strand exchange reactions with at least the same efficiency as the Escherichia coli SSB homotetramer.
Conclusions: The correct Deinococcus radiodurans ssb gene is a contiguous open reading frame that codes for the largest bacterial SSB monomer identified to date. The Deinococcus radiodurans SSB protein includes two OB folds per monomer and functions as a homodimer. The Deinococcus radiodurans SSB protein efficiently stimulates Deinococcus radiodurans RecA and also Escherichia coli RecA protein-promoted DNA strand exchange reactions. The identification and purification of Deinococcus radiodurans SSB protein not only allows for greater understanding of the SSB protein family but provides an essential yet previously missing player in the current efforts to understand the extraordinary DNA repair capacity of Deinococcus radiodurans.
Figures




References
-
- Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. - DOI - PMC - PubMed
-
- Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochemistry. 1994;63:527–570. - PubMed
-
- White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, Moffat KS, Qin HY, Jiang LX, Pamphile W, Crosby M, Shen M, Vamathevan JJ, Lam P, McDonald L, Utterback T, Zalewski C, Makarova KS, Aravind L, Daly MJ, Minton KW, Fraser CM, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. - DOI - PMC - PubMed
-
- Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore RJ, Romine MF, Shen Y, Stritmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci U S A. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

