Inter-species differences of co-expression of neighboring genes in eukaryotic genomes
- PMID: 14718066
- PMCID: PMC331401
- DOI: 10.1186/1471-2164-5-4
Inter-species differences of co-expression of neighboring genes in eukaryotic genomes
Abstract
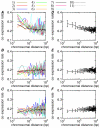
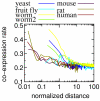
Background: There is increasing evidence that gene order within the eukaryotic genome is not random. In yeast and worm, adjacent or neighboring genes tend to be co-expressed. Clustering of co-expressed genes has been found in humans, worm and fruit flies. However, in mice and rats, an effect of chromosomal distance (CD) on co-expression has not been investigated yet. Also, no cross-species comparison has been made so far. We analyzed the effect of CD as well as normalized distance (ND) using expression data in six eukaryotic species: yeast, fruit fly, worm, rat, mouse and human.
Results: We analyzed 24 sets of expression data from the six species. Highly co-expressed pairs were sorted into bins of equal sized intervals of CD, and a co-expression rate (CoER) in each bin was calculated. In all datasets, a higher CoER was obtained in a short CD range than a long distance range. These results show that across all studied species, there was a consistent effect of CD on co-expression. However, the results using the ND show more diversity. Intra- and inter-species comparisons of CoER reveal that there are significant differences in the co-expression rates of neighboring genes among the species. A pair-wise BLAST analysis finds 8-30 % of the highly co-expressed pairs are duplicated genes.
Conclusion: We confirmed that in the six eukaryotic species, there was a consistent tendency that neighboring genes are likely to be co-expressed. Results of pair-wised BLAST indicate a significant effect of non-duplicated pairs on co-expression. A comparison of CD and ND suggests the dominant effect of CD.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

