Adenylyl cyclase G is activated by an intramolecular osmosensor
- PMID: 14718564
- PMCID: PMC363170
- DOI: 10.1091/mbc.e03-08-0622
Adenylyl cyclase G is activated by an intramolecular osmosensor
Abstract
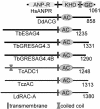
Adenylyl cyclase G (ACG) is activated by high osmolality and mediates inhibition of spore germination by this stress factor. The catalytic domains of all eukaryote cyclases are active as dimers and dimerization often mediates activation. To investigate the role of dimerization in ACG activation, we coexpressed ACG with an ACG construct that lacked the catalytic domain (ACGDeltacat) and was driven by a UV-inducible promoter. After UV induction of ACGDeltacat, cAMP production by ACG was strongly inhibited, but osmostimulation was not reduced. Size fractionation of native ACG showed that dimers were formed between ACG molecules and between ACG and ACGDeltacat. However, high osmolality did not alter the dimer/monomer ratio. This indicates that ACG activity requires dimerization via a region outside the catalytic domain but that dimer formation does not mediate activation by high osmolality. To establish whether ACG required auxiliary sensors for osmostimulation, we expressed ACG cDNA in a yeast adenylyl cyclase null mutant. In yeast, cAMP production by ACG was similarly activated by high osmolality as in Dictyostelium. This strongly suggests that the ACG osmosensor is intramolecular, which would define ACG as the first characterized primary osmosensor in eukaryotes.
Figures
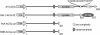





References
-
- Baldauf, S.L., Roger, A.J., Wenk-Siefert, I., and Doolittle, W.F. (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290, 972-977. - PubMed
-
- Bilwes, A.M., Alex, L.A., Crane, B.R., and Simon, M.I. (1999). Structure of CheA, a signal-transducing histidine kinase. Cell 96, 131-141. - PubMed
-
- Burke, D., Dawson, D., and Stearn, T. (2000). Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

