Evolution of cell recognition by viruses: a source of biological novelty with medical implications
- PMID: 14719364
- PMCID: PMC7119103
- DOI: 10.1016/s0065-3527(03)62002-6
Evolution of cell recognition by viruses: a source of biological novelty with medical implications
Abstract
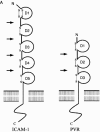
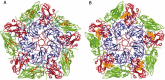
The picture beginning to form from genome analyses of viruses, unicellular organisms, and multicellular organisms is that viruses have shared functional modules with cells. A process of coevolution has probably involved exchanges of genetic information between cells and viruses for long evolutionary periods. From this point of view present-day viruses show flexibility in receptor usage and a capacity to alter through mutation their receptor recognition specificity. It is possible that for the complex DNA viruses, due to a likely limited tolerance to generalized high mutation rates, modifications in receptor specificity will be less frequent than for RNA viruses, albeit with similar biological consequences once they occur. It is found that different receptors, or allelic forms of one receptor, may be used with different efficiency and receptor affinities are probably modified by mutation and selection. Receptor abundance and its affinity for a virus may modulate not only the efficiency of infection, but also the capacity of the virus to diffuse toward other sites of the organism. The chapter concludes that receptors may be shared by different, unrelated viruses and that one virus may use several receptors and may expand its receptor specificity in ways that, at present, are largely unpredictable.
Figures



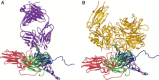
References
-
- Aderem A, Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. - PubMed
-
- Agace W.W, Amara A, Roberts A.I, Pablos J.L, Thelen S, Uguccioni M, Li X.Y, Marsal J, Arenzana-Seisdedos F, Delaunay T, Ebert E.C, Moser B, Parker C.M. Constitutive expression of stromal derived factor-1 by mucosal epithelia and its role in HIV transmission and propagation. Curr. Biol. 2000;10(6):325–328. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

