Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA
- PMID: 14722228
- PMCID: PMC373314
- DOI: 10.1093/nar/gnh015
Revised UV extinction coefficients for nucleoside-5'-monophosphates and unpaired DNA and RNA
Abstract
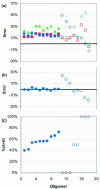
Ultraviolet absorption provides the nearly universal basis for determining concentrations of nucleic acids. Values for the UV extinction coefficients of DNA and RNA rely on the mononucleotide values determined 30-50 years ago. We show that nearly all of the previously published extinction coefficients for the nucleoside-5'-monophosphates are too large, and in error by as much as 7%. Concentrations based on complete hydrolysis and the older set of values are too low by approximately 4% for typical RNA and 2-3% for typical DNA samples. We also analyzed data in the literature for the extinction coefficients of unpaired DNA oligomers. Robust prediction of concentrations can be made using 38 microg/A260 unit for single-stranded DNA (ssDNA) having non-repetitive sequences and 40-80% GC. This is superior to currently used predictions that account for nearest-neighbor frequency or base composition. The latter result in concentrations that are 10-30% too low for typical ssDNA used as primers for PCR and other similar techniques. Methods are described here to accurately measure concentrations of nucleotides by nuclear magnetic resonance. NMR can be used to accurately determine concentrations (and extinction coefficients) of biomolecules within 1%.
Figures




References
-
- Cavaluzzi M.J., Kerwood,D.J. and Borer,P.N. (2002) Accurate nucleic acid concentrations by nuclear magnetic resonance. Anal. Biochem., 308, 373–380. - PubMed
-
- Cavaluzzi M.J. (2003) PhD dissertation, Syracuse University, NY, USA.
-
- Lide D.R. (2002) Handbook of Chemistry and Physics. CRC Press, Boca Raton, FL, USA.
-
- Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX PIPES. J. Biomol. NMR, 6, 277–293. - PubMed
-
- Gray D.M., Ratliff,R.L. and Vaughan,M.R. (1992) Circular dichroism spectroscopy of DNA. Methods Enzymol., 211, 389–406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

