Novel insect picorna-like virus identified in the brains of aggressive worker honeybees
- PMID: 14722264
- PMCID: PMC321398
- DOI: 10.1128/jvi.78.3.1093-1100.2004
Novel insect picorna-like virus identified in the brains of aggressive worker honeybees
Abstract
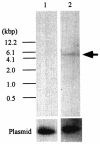
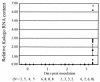
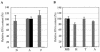
To identify candidate genes involved in the aggressive behavior of worker honeybees, we used the differential display method to search for RNAs exclusively detected in the brains of aggressive workers that had attacked a hornet. We identified a novel, 10,152-nucleotide RNA, termed Kakugo RNA. Kakugo RNA encodes a protein of 2,893 amino acid residues that shares structural features and sequence similarities with various picorna-like virus polyproteins, especially those from sacbrood virus, which infects honeybees. The Kakugo protein contains several domains that correspond to the virion protein, helicase, protease, and RNA-dependent RNA polymerase domains of various picorna-like virus polyproteins. When the worker bee tissue lysate was subjected to sucrose density gradient centrifugation, Kakugo RNA, except for the material at the bottom, was separated into two major peaks. One of the peaks corresponded to the position of Kakugo mRNA, and the other corresponded to the position of the poliovirus virion. These results suggest that the Kakugo RNA exists as an mRNA-like free RNA and virion RNA in the honeybee. Furthermore, injection of the lysate supernatant from the attacker heads into the heads of noninfected bees resulted in a marked increase in Kakugo RNA. These results demonstrate that Kakugo RNA is a plus-strand RNA of a novel picorna-like virus and that the brains of aggressive workers are infected by this novel virus. Kakugo RNA was detected in aggressive workers but not in nurse bees or foragers. In aggressive workers, Kakugo RNA was detected in the brain but not in the thorax or abdomen, indicating a close relation between viral infection in the brain and aggressive worker behaviors.
Figures




References
-
- Bailey, L. 1969. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 63:483-491. - PubMed
-
- Bailey, L., and E. F. W. Fernando. 1972. Effects of sacbrood virus on adult honey-bees. Ann. Appl. Biol. 72:27-35.
-
- Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 73:101-106. - PubMed
-
- Breed, M. D., G. E. Robinson, and J. R. E. Page. 1990. Division of labor during honey bee colony defense. Behav. Ecol. Sociobiol. 27:395-401.
-
- Breed, M. D., T. A. Smith, and A. Torres. 1992. Role of guard honey bees (Hymenoptera: Apidae) in nestmate discrimination and replacement of removed guards. Ann. Entomol. Soc. 85:633-637.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

