Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression
- PMID: 14722269
- PMCID: PMC321404
- DOI: 10.1128/jvi.78.3.1139-1149.2004
Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression
Abstract
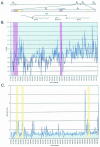
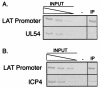
During herpes simplex virus type 1 (HSV-1) latency, gene expression is tightly repressed except for the latency-associated transcript (LAT). The mechanistic basis for this repression is unknown, but its global nature suggests regulation by an epigenetic mechanism such as DNA methylation. Previous work demonstrated that latent HSV-1 genomes are not extensively methylated, but these studies lacked the resolution to examine methylation of individual CpGs that could repress transcription from individual promoters during latency. To address this point, we employed established models to predict genomic regions with the highest probability of being methylated and, using bisulfite sequencing, analyzed the methylation profiles of these regions. We found no significant methylation of latent DNA isolated from mouse dorsal root ganglia in any of the regions examined, including the ICP4 and LAT promoters. This analysis indicates that methylation is unlikely to play a major role in regulating HSV-1 latent gene expression. Subsequently we focused on differential histone modification as another epigenetic mechanism that could regulate latent transcription. Chromatin immunoprecipitation analysis of the latent HSV-1 DNA repeat regions demonstrated that a portion of the LAT region is associated with histone H3 acetylated at lysines 9 and 14, consistent with a euchromatic and nonrepressed structure. In contrast, the chromatin associated with the HSV-1 DNA polymerase gene located in the unique long segment was not enriched in H3 acetylated at lysines 9 and 14, suggesting a transcriptionally inactive structure. These data suggest that histone composition may be a major regulatory determinant of HSV latency.
Figures


Similar articles
-
Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection.Proc Natl Acad Sci U S A. 2005 Nov 1;102(44):16055-9. doi: 10.1073/pnas.0505850102. Epub 2005 Oct 24. Proc Natl Acad Sci U S A. 2005. PMID: 16247011 Free PMC article.
-
The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription.J Virol. 2004 Nov;78(22):12508-18. doi: 10.1128/JVI.78.22.12508-12518.2004. J Virol. 2004. PMID: 15507638 Free PMC article.
-
A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities.J Virol. 2006 Mar;80(5):2358-68. doi: 10.1128/JVI.80.5.2358-2368.2006. J Virol. 2006. PMID: 16474142 Free PMC article.
-
Epigenotypes of latent herpesvirus genomes.Curr Top Microbiol Immunol. 2006;310:61-80. doi: 10.1007/3-540-31181-5_5. Curr Top Microbiol Immunol. 2006. PMID: 16909907 Review.
-
Epigenetic regulation of latent HSV-1 gene expression.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):246-56. doi: 10.1016/j.bbagrm.2009.12.001. Epub 2010 Jan 4. Biochim Biophys Acta. 2010. PMID: 20045093 Free PMC article. Review.
Cited by
-
Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3.Proc Natl Acad Sci U S A. 2013 Jul 9;110(28):E2621-8. doi: 10.1073/pnas.1309906110. Epub 2013 Jun 20. Proc Natl Acad Sci U S A. 2013. PMID: 23788661 Free PMC article.
-
The dynamics of HCF-1 modulation of herpes simplex virus chromatin during initiation of infection.Viruses. 2013 May 22;5(5):1272-91. doi: 10.3390/v5051272. Viruses. 2013. PMID: 23698399 Free PMC article. Review.
-
Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle.Interdiscip Perspect Infect Dis. 2010;2010:262415. doi: 10.1155/2010/262415. Epub 2010 Feb 15. Interdiscip Perspect Infect Dis. 2010. PMID: 20169002 Free PMC article.
-
Histone modification pattern of the T-cellular Herpesvirus saimiri genome in latency.J Virol. 2007 Mar;81(5):2524-30. doi: 10.1128/JVI.01931-06. Epub 2006 Dec 6. J Virol. 2007. PMID: 17151105 Free PMC article.
-
Tissue-specific splicing of the herpes simplex virus type 1 latency-associated transcript (LAT) intron in LAT transgenic mice.J Virol. 2006 Oct;80(19):9414-23. doi: 10.1128/JVI.00530-06. J Virol. 2006. PMID: 16973547 Free PMC article.
References
-
- Antequera, F., and A. Bird. 1993. CpG islands. EXS 64:169-185. - PubMed
-
- Baylin, S. B., J. G. Herman, J. R. Graff, P. M. Vertino, and J. P. Issa. 1998. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 72:141-196. - PubMed
-
- Bernstein, D. I., and J. C. Kappes. 1988. Enhanced in vitro reactivation of latent herpes simplex virus from neural and peripheral tissues with hexamethylenebisacetamide. Arch. Virol. 99:57-65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

