Quantification of the DNA cleavage and packaging proteins U(L)15 and U(L)28 in A and B capsids of herpes simplex virus type 1
- PMID: 14722291
- PMCID: PMC321391
- DOI: 10.1128/jvi.78.3.1367-1374.2004
Quantification of the DNA cleavage and packaging proteins U(L)15 and U(L)28 in A and B capsids of herpes simplex virus type 1
Abstract
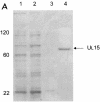
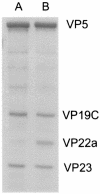
The proteins produced by the herpes simplex virus type 1 (HSV-1) genes U(L)15 and U(L)28 are believed to form part of the terminase enzyme, a protein complex essential for the cleavage of newly synthesized, concatameric herpesvirus DNA and the packaging of the resultant genome lengths into preformed capsids. This work describes the purification of recombinant forms of pU(L)15 and pU(L)28, which allowed the calculation of the average number of copies of each protein in A and B capsids and in capsids lacking the putative portal encoded by U(L)6. On average, 1.0 (+/-0.29 [standard deviation]) copies of pU(L)15 and 2.4 (+/-0.97) copies of pU(L)28 were present in B capsids, 1.2 (+/-0.72) copies of pU(L)15 and 1.5 (+/-0.86) copies of pU(L)28 were found in mutant capsids lacking the putative portal protein pU(L)6, and approximately 12.0 (+/-5.63) copies of pU(L)15 and 0.6 (+/-0.32) copies of pU(L)28 were present in each A capsid. These results suggest that the packaging machine is partly comprised of approximately 12 copies of pU(L)15, as found in A capsids, with wild-type B and mutant U(L)6(-) capsids containing an incomplete complement of cleavage and packaging proteins. These results are consistent with observations that B capsids form by default in the absence of packaging machinery in vitro and in vivo. In contrast, A capsids may be the result of initiated but aborted attempts at DNA packaging, resulting in the retention of at least part of the DNA packaging machinery.
Figures






Similar articles
-
The DNA cleavage and packaging protein encoded by the UL33 gene of herpes simplex virus 1 associates with capsids.Virology. 2004 Jul 1;324(2):475-82. doi: 10.1016/j.virol.2004.03.044. Virology. 2004. PMID: 15207632
-
The putative leucine zipper of the UL6-encoded portal protein of herpes simplex virus 1 is necessary for interaction with pUL15 and pUL28 and their association with capsids.J Virol. 2009 May;83(9):4557-64. doi: 10.1128/JVI.00026-09. Epub 2009 Feb 18. J Virol. 2009. PMID: 19224991 Free PMC article.
-
Role of the Herpes Simplex Virus CVSC Proteins at the Capsid Portal Vertex.J Virol. 2020 Nov 23;94(24):e01534-20. doi: 10.1128/JVI.01534-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967953 Free PMC article.
-
Nucleocapsid Assembly of Baculoviruses.Viruses. 2019 Jul 1;11(7):595. doi: 10.3390/v11070595. Viruses. 2019. PMID: 31266177 Free PMC article. Review.
-
The Terminase Complex of Each Human Herpesvirus.Biol Pharm Bull. 2024;47(5):912-916. doi: 10.1248/bpb.b23-00717. Biol Pharm Bull. 2024. PMID: 38692868 Review.
Cited by
-
Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides.J Virol. 2005 Dec;79(23):14660-7. doi: 10.1128/JVI.79.23.14660-14667.2005. J Virol. 2005. PMID: 16282466 Free PMC article.
-
A mutation in UL15 of herpes simplex virus 1 that reduces packaging of cleaved genomes.J Virol. 2011 Nov;85(22):11972-80. doi: 10.1128/JVI.00857-11. Epub 2011 Aug 31. J Virol. 2011. PMID: 21880766 Free PMC article.
-
Identification of a spliced gene from duck enteritis virus encoding a protein homologous to UL15 of herpes simplex virus 1.Virol J. 2011 Apr 6;8:156. doi: 10.1186/1743-422X-8-156. Virol J. 2011. PMID: 21466705 Free PMC article.
-
The putative herpes simplex virus 1 chaperone protein UL32 modulates disulfide bond formation during infection.J Virol. 2015 Jan;89(1):443-53. doi: 10.1128/JVI.01913-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320327 Free PMC article.
-
Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus.J Virol. 2007 Jun;81(12):6419-33. doi: 10.1128/JVI.00047-07. Epub 2007 Mar 28. J Virol. 2007. PMID: 17392365 Free PMC article.
References
-
- Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. - PubMed
-
- Addison, C., F. J. Rixon, J. W. Palfreyman, M. Ohara, and V. G. Preston. 1984. Characterization of a herpes simplex virus type-1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. - PubMed
-
- Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type-1 UL28 gene-product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. - PubMed
-
- Alkobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene-product is required for the assembly of full capsids. Virology 180:380-388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

