CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations
- PMID: 14722619
- PMCID: PMC311435
- DOI: 10.1172/JCI19874
CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations
Abstract
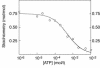
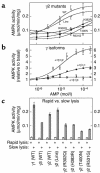
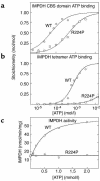
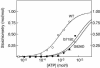
CBS domains are defined as sequence motifs that occur in several different proteins in all kingdoms of life. Although thought to be regulatory, their exact functions have been unknown. However, their importance was underlined by findings that mutations in conserved residues within them cause a variety of human hereditary diseases, including (with the gene mutated in parentheses): Wolff-Parkinson-White syndrome (gamma 2 subunit of AMP-activated protein kinase); retinitis pigmentosa (IMP dehydrogenase-1); congenital myotonia, idiopathic generalized epilepsy, hypercalciuric nephrolithiasis, and classic Bartter syndrome (CLC chloride channel family members); and homocystinuria (cystathionine beta-synthase). AMP-activated protein kinase is a sensor of cellular energy status that is activated by AMP and inhibited by ATP, but the location of the regulatory nucleotide-binding sites (which are prime targets for drugs to treat obesity and diabetes) was not characterized. We now show that tandem pairs of CBS domains from AMP-activated protein kinase, IMP dehydrogenase-2, the chloride channel CLC2, and cystathionine beta-synthase bind AMP, ATP, or S-adenosyl methionine,while mutations that cause hereditary diseases impair this binding. This shows that tandem pairs of CBS domains act, in most cases, as sensors of cellular energy status and, as such, represent a newly identified class of binding domain for adenosine derivatives.
Figures



Comment in
-
Bateman domains and adenosine derivatives form a binding contract.J Clin Invest. 2004 Jan;113(2):182-4. doi: 10.1172/JCI20846. J Clin Invest. 2004. PMID: 14722609 Free PMC article. Review.
References
-
- Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997;22:12–13. - PubMed
-
- Kennan A, et al. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wild-type and Rho(–/–) mice. Hum. Mol. Genet. 2002;11:547–557. - PubMed
-
- Pusch M. Myotonia caused by mutations in the muscle chloride channel gene CLCN1. Hum. Mutat. 2002;19:423–434. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

