Calorie restriction extends yeast life span by lowering the level of NADH
- PMID: 14724176
- PMCID: PMC314267
- DOI: 10.1101/gad.1164804
Calorie restriction extends yeast life span by lowering the level of NADH
Abstract
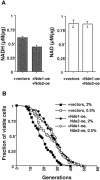
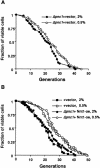
Calorie restriction (CR) extends life span in a wide variety of species. Previously, we showed that calorie restriction increases the replicative life span in yeast by activating Sir2, a highly conserved NAD-dependent deacetylase. Here we test whether CR activates Sir2 by increasing the NAD/NADH ratio or by regulating the level of nicotinamide, a known inhibitor of Sir2. We show that CR decreases NADH levels, and that NADH is a competitive inhibitor of Sir2. A genetic intervention that specifically decreases NADH levels increases life span, validating the model that NADH regulates yeast longevity in response to CR.
Figures


References
-
- Anderson R.M., Bitterman, K.J., Wood, J.G., Medvedik, O., Cohen, H., Lin, S.S., Manchester, J.K., Gordon, J.I., and Sinclair, D.A. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277: 18881–18890. - PubMed
-
- Bakker B.M., Overkamp, K.M., van Maris, A.J., Kotter, P., Luttik, M.A., van Dijken, J.P., and Pronk, J.T. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25: 15–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
