Expression array annotation using the BioMediator biological data integration system and the BioConductor analytic platform
- PMID: 14728212
- PMCID: PMC1479993
Expression array annotation using the BioMediator biological data integration system and the BioConductor analytic platform
Abstract
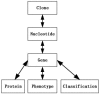
This paper presents the implementation of a model for expression array annotation (EAA) using the BioMediator biological data integration system along with BioConductor, an analytic tools platform. The model presented addresses the need for annotation sources identified during BioConductor inverted exclamation mark s development. Annotation provides us with well-curated genomic background knowledge for expression array analysis and interpretation. Annotation requests are constructed and posted to the query interface of the EAA package (the EAA model implemented as a component of BioConductor). The software enumerates all possible annotation paths for queries. These are then transformed to PQL queries and processed by BioMediator. Annotation entities returned from the EAA package answer the annotation request.
Figures
References
-
- Gene Ontology Consortium (GO). Gene ontology (go). http://www.geneontology.org/
-
- Hvidsten TR, Komorowski J, Sandvik AK, Laegreid A. Predicting gene function from gene expressions and ontologies. In: Pacific Symposium on Biocomputing; 2001; 2001. p. 299–310. - PubMed
-
- Bioconductor. http://www.bioconductor.org/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources