Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12
- PMID: 14728724
- PMCID: PMC343271
- DOI: 10.1186/1471-2164-5-8
Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12
Abstract
Background: The human 6-16 and ISG12 genes are transcriptionally upregulated in a variety of cell types in response to type I interferon (IFN). The predicted products of these genes are small (12.9 and 11.5 kDa respectively), hydrophobic proteins that share 36% overall amino acid identity. Gene disruption and over-expression studies have so far failed to reveal any biochemical or cellular roles for these proteins.
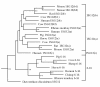
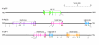
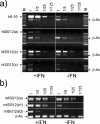
Results: We have used in silico analyses to identify a novel family of genes (the ISG12 gene family) related to both the human 6-16 and ISG12 genes. Each ISG12 family member codes for a small hydrophobic protein containing a conserved ~80 amino-acid motif (the ISG12 motif). So far we have detected 46 family members in 25 organisms, ranging from unicellular eukaryotes to humans. Humans have four ISG12 genes: the 6-16 gene at chromosome 1p35 and three genes (ISG12(a), ISG12(b) and ISG12(c)) clustered at chromosome 14q32. Mice have three family members (ISG12(a), ISG12(b1) and ISG12(b2)) clustered at chromosome 12F1 (syntenic with human chromosome 14q32). There does not appear to be a murine 6-16 gene. On the basis of phylogenetic analyses, genomic organisation and intron-alignments we suggest that this family has arisen through divergent inter- and intra-chromosomal gene duplication events. The transcripts from human and mouse genes are detectable, all but two (human ISG12(b) and ISG12(c)) being upregulated in response to type I IFN in the cell lines tested.
Conclusions: Members of the eukaryotic ISG12 gene family encode a small hydrophobic protein with at least one copy of a newly defined motif of approximately 80 amino-acids (the ISG12 motif). In higher eukaryotes, many of the genes have acquired a responsiveness to type I IFN during evolution suggesting that a role in resisting cellular or environmental stress may be a unifying property of all family members. Analysis of gene-function in higher eukaryotes is complicated by the possibility of functional redundancy between family-members. Genetic studies in organisms (e.g. Dictyostelium discoideum) with just one family member so far identified may be particularly helpful in this respect.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

