In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database
- PMID: 14729914
- PMCID: PMC341918
- DOI: 10.1105/tpc.017814
In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database
Abstract
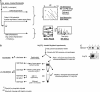
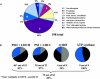
An extensive analysis of the Arabidopsis thaliana peripheral and integral thylakoid membrane proteome was performed by sequential extractions with salt, detergent, and organic solvents, followed by multidimensional protein separation steps (reverse-phase HPLC and one- and two-dimensional electrophoresis gels), different enzymatic and nonenzymatic protein cleavage techniques, mass spectrometry, and bioinformatics. Altogether, 154 proteins were identified, of which 76 (49%) were alpha-helical integral membrane proteins. Twenty-seven new proteins without known function but with predicted chloroplast transit peptides were identified, of which 17 (63%) are integral membrane proteins. These new proteins, likely important in thylakoid biogenesis, include two rubredoxins, a potential metallochaperone, and a new DnaJ-like protein. The data were integrated with our analysis of the lumenal-enriched proteome. We identified 83 out of 100 known proteins of the thylakoid localized photosynthetic apparatus, including several new paralogues and some 20 proteins involved in protein insertion, assembly, folding, or proteolysis. An additional 16 proteins are involved in translation, demonstrating that the thylakoid membrane surface is an important site for protein synthesis. The high coverage of the photosynthetic apparatus and the identification of known hydrophobic proteins with low expression levels, such as cpSecE, Ohp1, and Ohp2, indicate an excellent dynamic resolution of the analysis. The sequential extraction process proved very helpful to validate transmembrane prediction. Our data also were cross-correlated to chloroplast subproteome analyses by other laboratories. All data are deposited in a new curated plastid proteome database (PPDB) with multiple search functions (http://cbsusrv01.tc.cornell.edu/users/ppdb/). This PPDB will serve as an expandable resource for the plant community.
Figures






References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Aro, E.-M., and Andersson, B. (2001). Regulation of Photosynthesis. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

