MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana
- PMID: 14729917
- PMCID: PMC341911
- DOI: 10.1105/tpc.018630
MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana
Abstract
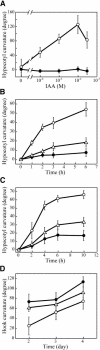
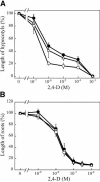
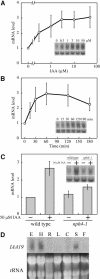
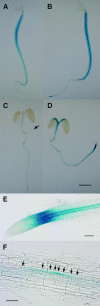
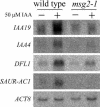
We have isolated a dominant, auxin-insensitive mutant of Arabidopsis thaliana, massugu2 (msg2), that displays neither hypocotyl gravitropism nor phototropism, fails to maintain an apical hook as an etiolated seedling, and is defective in lateral root formation. Yet other aspects of growth and development of msg2 plants are almost normal. These characteristics of msg2 are similar to those of another auxin-insensitive mutant, non-phototropic hypocotyl4 (nph4), which is a loss-of-function mutant of AUXIN RESPONSE FACTOR7 (ARF7) (Harper et al., 2000). Map-based cloning of the MSG2 locus reveals that all four mutant alleles result in amino acid substitutions in the conserved domain II of an Auxin/Indole-3-Acetic Acid protein, IAA19. Interestingly, auxin inducibility of MSG2/IAA19 gene expression is reduced by 65% in nph4/arf7. Moreover, MSG2/IAA19 protein binds to the C-terminal domain of NPH4/ARF7 in a Saccharomyces cerevisiae (yeast) two-hybrid assay and to the whole latter protein in vitro by pull-down assay. These results suggest that MSG2/IAA19 and NPH4/ARF7 may constitute a negative feedback loop to regulate differential growth responses of hypocotyls and lateral root formation.
Figures





References
-
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. - PubMed
-
- Ainley, W.M., Walker, J.C., Nagao, R.T., and Key, J.L. (1988). Sequence and characterization of two auxin-regulated genes from soybean. J. Biol. Chem. 263, 10658–10666. - PubMed
-
- The Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1990). Current Protocols in Molecular Biology. (New York: Wiley InterScience).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

