RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export
- PMID: 14729961
- PMCID: PMC321439
- DOI: 10.1128/MCB.24.3.1155-1167.2004
RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export
Abstract
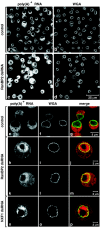
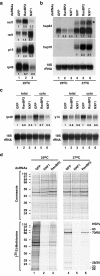
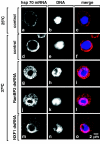
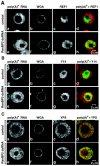
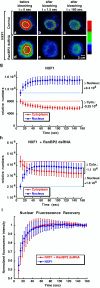
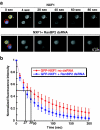
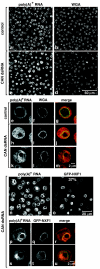
Metazoan NXF1-p15 heterodimers promote the nuclear export of bulk mRNA across nuclear pore complexes (NPCs). In vitro, NXF1-p15 forms a stable complex with the nucleoporin RanBP2/Nup358, a component of the cytoplasmic filaments of the NPC, suggesting a role for this nucleoporin in mRNA export. We show that depletion of RanBP2 from Drosophila cells inhibits proliferation and mRNA export. Concomitantly, the localization of NXF1 at the NPC is strongly reduced and a significant fraction of this normally nuclear protein is detected in the cytoplasm. Under the same conditions, the steady-state subcellular localization of other nuclear or cytoplasmic proteins and CRM1-mediated protein export are not detectably affected, indicating that the release of NXF1 into the cytoplasm and the inhibition of mRNA export are not due to a general defect in NPC function. The specific role of RanBP2 in the recruitment of NXF1 to the NPC is highlighted by the observation that depletion of CAN/Nup214 also inhibits cell proliferation and mRNA export but does not affect NXF1 localization. Our results indicate that RanBP2 provides a major binding site for NXF1 at the cytoplasmic filaments of the NPC, thereby restricting its diffusion in the cytoplasm after NPC translocation. In RanBP2-depleted cells, NXF1 diffuses freely through the cytoplasm. Consequently, the nuclear levels of the protein decrease and export of bulk mRNA is impaired.
Figures



References
-
- Bachi, A., I. C. Braun, J. P. Rodrigues, N. Panté, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Görlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. - PMC - PubMed
-
- Braun, I. C., A. Herold, M. Rode, E. Conti, and E. Izaurralde. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536-20543. - PubMed
-
- Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 3:310-319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
