Gene trapping with firefly luciferase in Arabidopsis. Tagging of stress-responsive genes
- PMID: 14730060
- PMCID: PMC316285
- DOI: 10.1104/pp.103.027151
Gene trapping with firefly luciferase in Arabidopsis. Tagging of stress-responsive genes
Abstract
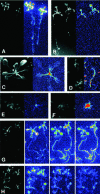
To monitor the expression of T-DNA-tagged plant genes in vivo, a collection of 20,261 transgenic lines of Arabidopsis (Columbia-0) were generated with the promoter trap vector pTluc, which carries a promoterless firefly luc (luciferase) reporter gene linked to the right T-DNA border. By detection of bioluminescence in 3-week-old seedlings, 753 lines were identified showing constitutive, organ-specific, and stress-responsive luciferase expression patterns. To facilitate the identification of well-defined luciferase expression patterns, a pooled seed stock was established. Several lines showed sugar, salt, and abscisic acid (ABA)-inducible luciferase activity. Segregation analysis of 215 promoter trap lines indicated that about 50% of plants contained single insertions, whereas 40% carried two and 10% carried three or more T-DNA tags. Sequencing the T-DNA insert junctions isolated from 17 luciferase-expressing lines identified T-DNA tags in 5'- and 3'-transcribed domains and translational gene fusions generated by T-DNA insertions in exons and introns of Arabidopsis genes. Tissue specific expression of eight wild-type Arabidopsis genes was confirmed to be similar to the luminescence patterns observed in the corresponding luciferase-tagged lines. Here, we describe the characterization of a transcriptional luc reporter gene fusion with the WBC-type ABC transporter gene At1g17840. Expression of wild-type and luciferase-tagged At1g17840 alleles revealed similar induction by salt, glucose, and ABA treatments and gibberellin-mediated down-regulation of ABA-induced expression. These results illustrate that luciferase gene traps are well suited for monitoring the expression of stress-responsive Arabidopsis genes in vivo.
Figures







References
-
- Allen ND, Cran DG, Barton SC, Hettle S, Reik W, Surani MA (1988) Transgenes as probes for active chromosomal domains in mouse development. Nature 333: 852-855 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 - PubMed
-
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259-266 - PubMed
-
- Bellen HJ, O'Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ (1989) P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev 3: 1288-1300 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

