Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites
- PMID: 14732688
- PMCID: PMC327130
- DOI: 10.1073/pnas.0307540100
Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites
Abstract
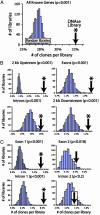
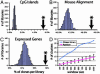
Analysis of the human genome sequence has identified approximately 25000-30000 protein-coding genes, but little is known about how most of these are regulated. Mapping DNase I hypersensitive (HS) sites has traditionally represented the gold-standard experimental method for identifying regulatory elements, but the labor-intensive nature of this technique has limited its application to only a small number of human genes. We have developed a protocol to generate a genome-wide library of gene regulatory sequences by cloning DNase HS sites. We generated a library of DNase HS sites from quiescent primary human CD4(+) T cells and analyzed approximately 5600 of the resulting clones. Compared to sequences from randomly generated in silico libraries, sequences from these clones were found to map more frequently to regions of the genome known to contain regulatory elements, such as regions upstream of genes, within CpG islands, and in sequences that align between mouse and human. These cloned sites also tend to map near genes that have detectable transcripts in CD4(+) T cells, demonstrating that transcriptionally active regions of the genome are being selected. Validation of putative regulatory elements was achieved by repeated recovery of the same sequence and real-time PCR. This cloning strategy, which can be scaled up and applied to any cell line or tissue, will be useful in identifying regulatory elements controlling global expression differences that delineate tissue types, stages of development, and disease susceptibility.
Figures



References
-
- Collins, F. S., Green, E. D., Guttmacher, A. E. & Guyer, M. S. (2003) Nature 422, 835–847. - PubMed
-
- Ureta-Vidal, A., Ettwiller, L. & Birney, E. (2003) Nat. Rev. Genet. 4, 251–262. - PubMed
-
- Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788–793. - PubMed
-
- Slightom, J. L., Bock, J. H., Tagle, D. A., Gumucio, D. L., Goodman, M., Stojanovic, N., Jackson, J., Miller, W. & Hardison, R. (1997) Genomics 39, 90–94. - PubMed
-
- Lieb, J. D., Liu, X., Botstein, D. & Brown, P. O. (2001) Nat. Genet. 28, 327–334. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

